Nomenclature
Short Name:
PFTAIRE2
Full Name:
ALS2CR7
Alias:
- A2S7
- EC 2.7.11.22
- PFTAIRE protein kinase 2
- PFTK2
- Serine/threonine-protein kinase ALS2CR7
- AL2S7
- ALS2CR7
- Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 7 protein
- Amyotrophic lateral sclerosis 2 chromosomal region candidate gene protein 7
- CDK15
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
CDK
SubFamily:
TAIRE
Structure
Mol. Mass (Da):
49,023
# Amino Acids:
435
# mRNA Isoforms:
5
mRNA Isoforms:
49,023 Da (435 AA; Q96Q40); 48,316 Da (429 AA; Q96Q40-5); 45,046 Da (400 AA; Q96Q40-3); 43,574 Da (384 AA; Q96Q40-4); 40,122 Da (358 AA; Q96Q40-2)
4D Structure:
NA
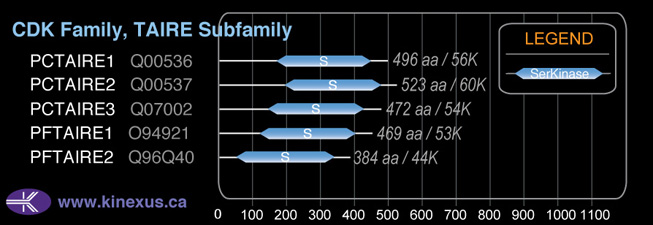
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 103 | 387 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S77.
Threonine phosphorylated:
T170.
Tyrosine phosphorylated:
Y114, Y118.
Ubiquitinated:
K239.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 72
72
1271
18
1340
 0.3
0.3
6
6
5
 -
-
-
-
-
 1.1
1.1
19
76
25
 29
29
517
27
299
 0.5
0.5
9
27
10
 0.3
0.3
6
33
6
 0.2
0.2
4
5
5
 0.1
0.1
2
3
0
 3
3
52
29
49
 0.3
0.3
6
5
7
 53
53
939
23
522
 -
-
-
-
-
 0.2
0.2
4
3
2
 0.3
0.3
6
5
5
 0.5
0.5
8
15
5
 0.8
0.8
14
92
11
 0.1
0.1
1
3
0
 3
3
59
21
43
 24
24
433
77
198
 0.2
0.2
3
5
3
 0.2
0.2
3
5
2
 -
-
-
-
-
 1.2
1.2
21
3
1
 0.2
0.2
4
5
4
 76
76
1350
53
3308
 0.1
0.1
2
3
1
 0.1
0.1
2
3
0
 0.1
0.1
2
3
1
 10
10
171
14
120
 26
26
459
12
38
 100
100
1768
21
3762
 0.1
0.1
2
36
0
 45
45
796
78
700
 6
6
112
48
124
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 91
91
91.3
99 58.3
58.3
59.6
97 -
-
-
91 -
-
-
- 92
92
94.8
92.5 -
-
-
- 89.4
89.4
92.9
91 41.7
41.7
56.6
86 -
-
-
- -
-
-
- 63.8
63.8
73.9
69 54.3
54.3
68
- 61.4
61.4
72.9
67 -
-
-
- 35.2
35.2
46.4
- 46.8
46.8
59.7
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 33.1
33.1
45.8
- 36.5
36.5
48.5
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CDK2 - P24941 |
| 2 | CENPK - Q9BS16 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| BIRC5 (Survivin) | O15392 | T34 | FLEGCACTPERMAEA | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
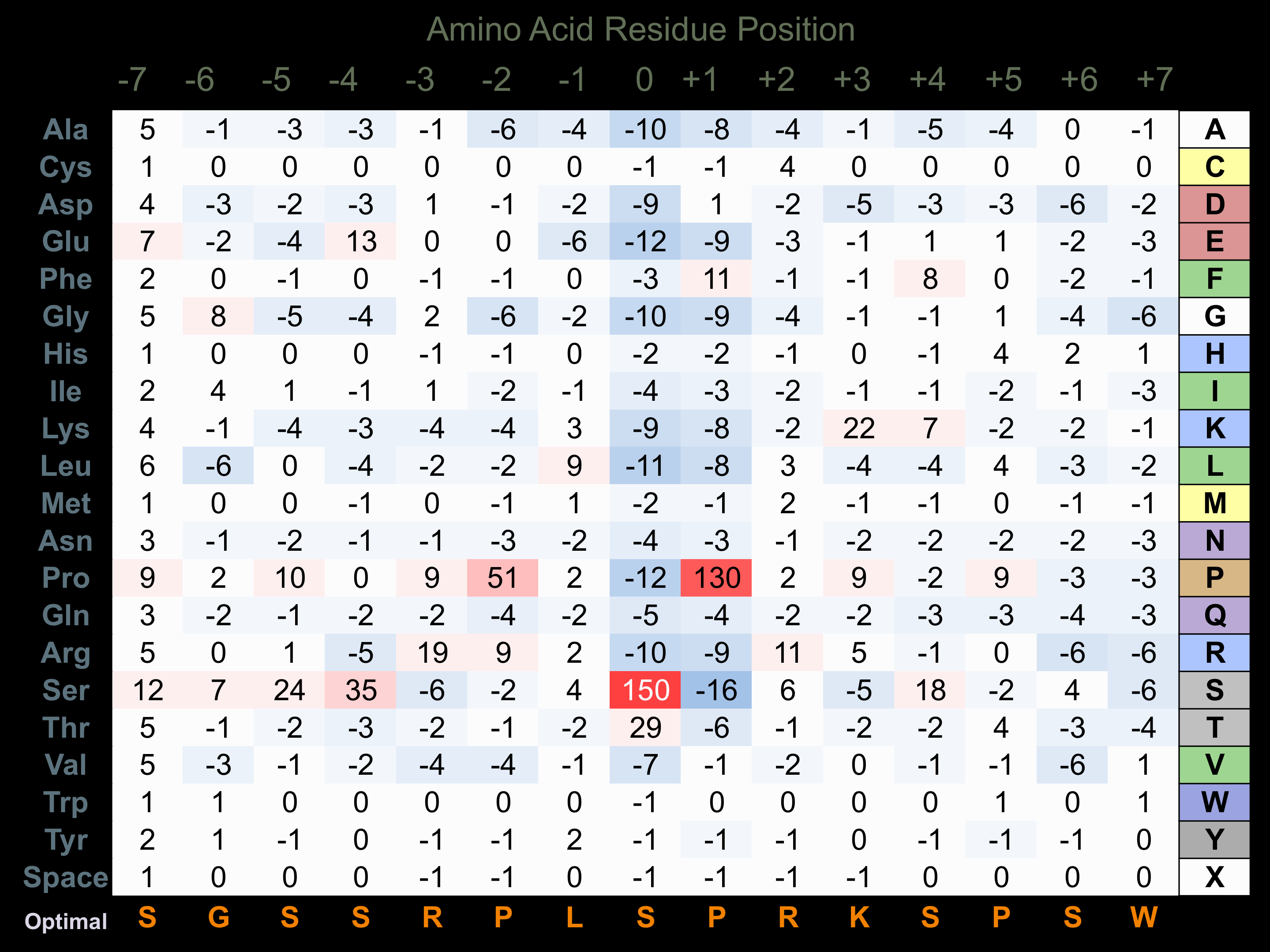
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| R547 | Kd = 7.2 nM | 6918852 | 22037378 | |
| Foretinib | Kd = 32 nM | 42642645 | 1230609 | 22037378 |
| AT7519 | Kd = 62 nM | 11338033 | 22037378 | |
| AST-487 | Kd = 84 nM | 11409972 | 574738 | 22037378 |
| Staurosporine | Kd = 260 nM | 5279 | 22037378 | |
| Alvocidib | Kd = 330 nM | 9910986 | 428690 | 22037378 |
| PHA-665752 | Kd = 510 nM | 10461815 | 450786 | 22037378 |
| AC1NS7CD | Kd = 740 nM | 5329665 | 295136 | 22037378 |
| SNS032 | Kd = 740 nM | 3025986 | 296468 | 22037378 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| Lestaurtinib | Kd = 1.3 µM | 126565 | 22037378 | |
| Bosutinib | Kd = 1.6 µM | 5328940 | 288441 | 22037378 |
| A674563 | Kd = 2.1 µM | 11314340 | 379218 | 22037378 |
| NVP-TAE684 | Kd = 2.1 µM | 16038120 | 509032 | 22037378 |
| KW2449 | Kd = 2.6 µM | 11427553 | 1908397 | 22037378 |
| Sorafenib | Kd = 2.9 µM | 216239 | 1336 | 19654408 |
| BMS-690514 | Kd < 3 µM | 11349170 | 21531814 | |
| Enzastaurin | Kd = 4.9 µM | 176167 | 300138 | 22037378 |
Disease Linkage
General Disease Association:
Neurological disorders
Specific Diseases (Non-cancerous):
Amyotrophic lateral sclerosis (ALS); Primary lateral sclerosis (PLS)
Comments:
ALS is a rare neuronal disease affecting motor control. PFTAIRE2 may be associated with amyotrophic lateral sclerosis 2 (ALS2), which is an autosomal recessive form of juvenile ALS. Characteristics of ALS can include cramping, stiffness, muscle twitching, weakness, slurring words, chewing difficulties, and dysphagia (difficulty swallowing). Lateral Sclerosis (PLS) is a rare neuronal disease related to the Amyotrophic Lateral Sclerosis disorder.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for PFTAIRE2 in diverse human cancers of 300, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25141 diverse cancer specimens. This rate is very similar (+ 6% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.34 % in 1270 large intestine cancers tested; 0.31 % in 1634 lung cancers tested; 0.29 % in 548 urinary tract cancers tested; 0.27 % in 603 endometrium cancers tested; 0.16 % in 864 skin cancers tested; 0.16 % in 589 stomach cancers tested; 0.12 % in 1316 breast cancers tested; 0.09 % in 1512 liver cancers tested; 0.09 % in 1276 kidney cancers tested; 0.04 % in 558 thyroid cancers tested; 0.03 % in 833 ovary cancers tested; 0.02 % in 1459 pancreas cancers tested; 0.01 % in 2082 central nervous system cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: V210M (3).
Comments:
Only 3 deletions, and no insertions or complex mutations are noted on the COSMIC website.

