Nomenclature
Short Name:
PIM1
Full Name:
Proto-oncogene serine-threonine-protein kinase Pim-1
Alias:
- EC 2.7.11.1
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
PIM
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
45,412
# Amino Acids:
404
# mRNA Isoforms:
2
mRNA Isoforms:
45,412 Da (404 AA; P11309); 35,686 Da (313 AA; P11309-2)
4D Structure:
Binds to RP9. Isoform 2 is isolated as a monomer whereas isoform 1 complexes with other proteins. Isoform 1, but not isoform 2, binds BMX. Isoform 2 interacts with CDKN1B and FOXO3.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 129 | 381 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S95, S99, S137, S189, S280+, S299-, S352.
Threonine phosphorylated:
T114+, T290+, T295-.
Tyrosine phosphorylated:
Y309+.
Ubiquitinated:
K96, K158, K260, K274, K285.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 13
13
1268
16
1646
 1.4
1.4
135
12
159
 1.2
1.2
113
5
112
 14
14
1292
67
2746
 8
8
746
14
683
 15
15
1420
51
3068
 7
7
623
23
784
 25
25
2313
38
3519
 5
5
442
10
350
 3
3
283
64
256
 2
2
232
21
318
 7
7
655
143
546
 2
2
147
17
76
 5
5
478
12
368
 2
2
219
15
252
 1.2
1.2
113
9
88
 3
3
294
192
153
 2
2
151
14
263
 1.2
1.2
116
54
73
 8
8
749
56
669
 2
2
188
19
158
 3
3
247
21
225
 3
3
311
23
281
 1.1
1.1
103
15
84
 2
2
194
19
179
 39
39
3655
44
5176
 2
2
167
20
95
 2
2
161
15
140
 3
3
239
15
229
 54
54
5116
14
1507
 18
18
1690
18
876
 100
100
9422
27
14618
 1.5
1.5
138
53
303
 9
9
872
36
745
 3
3
265
22
475
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 46.1
46.1
58
99.5 58.7
58.7
59.9
98 -
-
-
99 -
-
-
- 46.2
46.2
57.5
99 -
-
-
- 85.9
85.9
88.6
94 75.3
75.3
76.7
97 -
-
-
- 48
48
61.1
- -
-
-
83 51.5
51.5
65.1
74 37.4
37.4
51.5
73 -
-
-
- -
-
-
- -
-
-
- -
-
-
46 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CDC25A - P30304 |
| 2 | BAD - Q92934 |
| 3 | CBX1 - P83916 |
| 4 | PTPRO - Q16827 |
| 5 | ABCG2 - Q9UNQ0 |
| 6 | NFATC1 - O95644 |
| 7 | STAT3 - P40763 |
Regulation
Activation:
Phosphorylation of Ser-280 and Tyr-309 increases phosphotransferase activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ABCG2 | Q9UNQ0 | T362 | GEKKKKITVFKEISY | |
| AML2 | Q13761 | S149 | GRSGRGKSFTLTITV | |
| AML2 | Q13761 | T151 | SGRGKSFTLTITVFT | |
| AML2 | Q13761 | T153 | RGKSFTLTITVFTNP | |
| AML2 | Q13761 | T155 | KSFTLTITVFTNPTQ | |
| Bad | Q92934 | S118 | GRELRRMSDEFVDSF | - |
| Bad | Q92934 | S75 | EIRSRHSSYPAGTED | - |
| Bad | Q92934 | S99 | PFRGRSRSAPPNLWA | - |
| Cip1 (p21, CDKN1A) | P38936 | S145 | GRKRRQTSMTDFYHS | |
| Cip1 (p21, CDKN1A) | P38936 | T144 | QGRKRRQTSMTDFYH | - |
| FOXO3 (FKHRL1) | O43524 | S253 | APRRRAVSMDNSNKY | - |
| FOXO3 (FKHRL1) | O43524 | T32 | QSRPRSCTWPLQRPE | |
| H3.1 | P68431 | S11 | TKQTARKSTGGKAPR | + |
| MARK3 | P27448 | S96 | KTQLNPTSLQKLFRE | - |
| MARK3 | P27448 | T90 | AIKIIDKTQLNPTSL | - |
| MARK3 | P27448 | T95 | DKTQLNPTSLQKLFR | - |
| MDM2 | Q00987 | S166 | SSRRRAISETEENSD | |
| MDM2 | Q00987 | S186 | RQRKRHKSDSISLSF | |
| Myc | P01106 | S329 | AKRVKLDSVRVLRQI | |
| p27Kip1 | P46527 | T157 | GIRKRPATDDSSTQN | - |
| p27Kip1 | P46527 | T198 | PGLRRRQT_______ | - |
| Pim1 | P11309 | S280 | LKLIDFGSGALLKDT | + |
| Pim1 | P11309 | S95 | LEVGMLLSKINSLAH | |
| Pim1 | P11309 | T295 | VYTDFDGTRVYSPPE | - |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
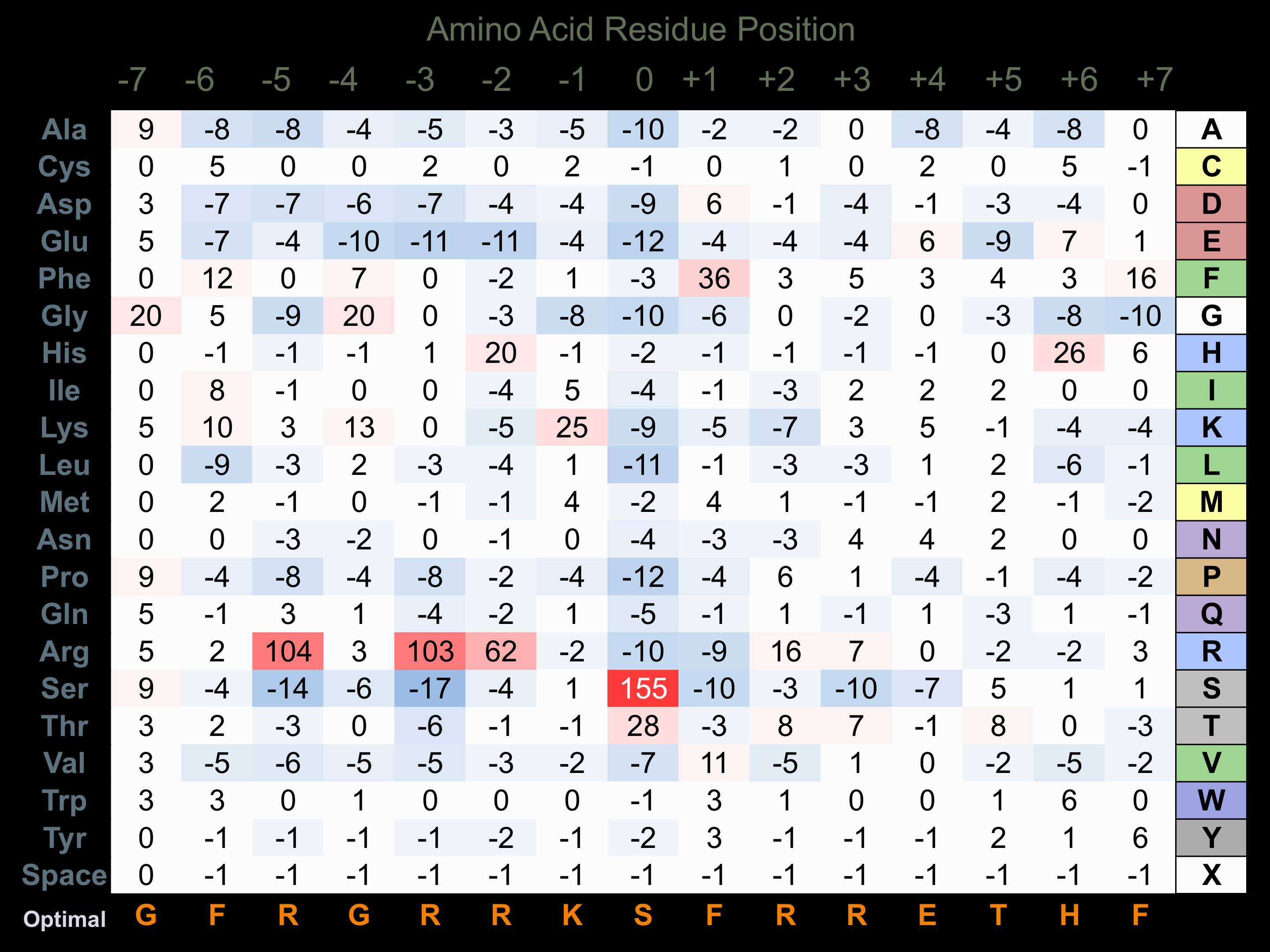
Matrix Type:
Experimentally derived from alignment of 33 known protein substrate phosphosites and 18 peptides phosphorylated by recombinant Pim1 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Prostate cancer; Marginal zone B-cell lymphomas; Nodal marginal zone B-Cell lymphomas; Lung cancer; Diffuse large B-cell lymphomas (DLBCL)
Comments:
PIM1 is a known oncoprotein (OP). The Pim1 protein is undergoes relatively little mutation in human tumours, about the same as typical proteins. The active form of the protein kinase normally acts to promote tumour cell proliferation. The oncogenic activity of the Pim-1 protein is mediated through the positive regulation of MYC transcription (another oncogene), the regulation of cell cycle progression, and by the inhibition of the pro-apoptotic proteins BAD, MAP3K5, and FOXO3. In addition, Pim-1 has been shown to promote genomic instability. Gain-of-function mutations in the Pim-1 gene resulting in the up-regulation of Pim-1 activity have been observed in several human cancer types, indicating a role in tumorigenesis. For example, significantly elevated Pim-1 expression was observed in several hematopoietic malignancies and in several solid tumours. In animal studies, over-expression of the Pim-1 protein in mice leads to an increased occurence of tumours, while the knockout of Pim-1 expression has no observable phenotypic consequence.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= -61, p<0.008); Cervical epithelial cancer (%CFC= +98, p<0.006); Classical Hodgkin lymphomas (%CFC= +49, p<0.002); Colorectal adenocarcinomas (early onset) (%CFC= +65, p<0.004); Large B-cell lymphomas (%CFC= +101, p<0.007); Oral squamous cell carcinomas (OSCC) (%CFC= -64, p<0.05); Skin fibrosarcomas (%CFC= -61); Skin melanomas - malignant (%CFC= -63, p<0.006); Skin squamous cell carcinomas (%CFC= +168, p<0.093); Vulvar intraepithelial neoplasia (%CFC= +58, p<0.035). The COSMIC website notes an up-regulated expression score for PIM1 in diverse human cancers of 418, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 2 for this protein kinase in human cancers was 97% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 26039 diverse cancer specimens. This rate is only 20 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.55 % in 2221 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: K24N (6); S97N (6).
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.

