Nomenclature
Short Name:
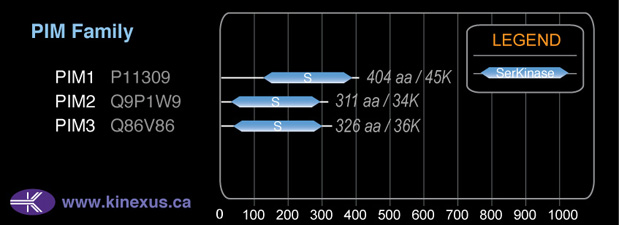
PIM2
Full Name:
Serine-threonine-protein kinase Pim-2
Alias:
- EC 2.7.11.1
- Pim-2h
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
PIM
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
34,190
# Amino Acids:
311
# mRNA Isoforms:
1
mRNA Isoforms:
34,190 Da (311 AA; Q9P1W9)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 32 | 286 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S185+, S204-.
Threonine phosphorylated:
T3, T15, T195+, T200-.
Tyrosine phosphorylated:
Y32.
Ubiquitinated:
K40, K61, K89, K132, K165, K179.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 28
28
603
19
1085
 2
2
51
12
31
 4
4
77
5
41
 38
38
824
55
2026
 28
28
604
14
591
 67
67
1438
45
3182
 24
24
524
23
673
 100
100
2156
30
5009
 15
15
332
10
259
 5
5
103
64
95
 3
3
70
22
59
 23
23
503
133
603
 13
13
282
17
174
 2
2
53
12
30
 2
2
52
15
47
 3
3
60
9
29
 5
5
113
191
500
 4
4
81
15
45
 1.4
1.4
31
54
20
 16
16
346
56
376
 3
3
66
19
42
 15
15
323
21
291
 8
8
180
15
102
 3
3
71
15
69
 8
8
171
19
191
 87
87
1865
35
4023
 9
9
188
20
106
 6
6
138
15
94
 3
3
64
15
47
 7
7
142
14
84
 70
70
1500
18
782
 27
27
592
21
654
 4
4
79
56
131
 35
35
746
31
756
 3
3
55
22
42
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 70.5
70.5
70.5
100 98.7
98.7
99
99 -
-
-
92 -
-
-
- 71.6
71.6
73.6
94 -
-
-
- 75.1
75.1
78.1
89 55.8
55.8
67.8
90 -
-
-
- 47.8
47.8
57.7
- -
-
-
- 49.2
49.2
63.2
- 52.4
52.4
67.8
- -
-
-
- -
-
-
- -
-
-
- -
-
-
37 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | BAD - Q92934 |
| 2 | ATXN1 - P54253 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
Down-regulated in response to enterovirus 71 (EV71) infection.
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
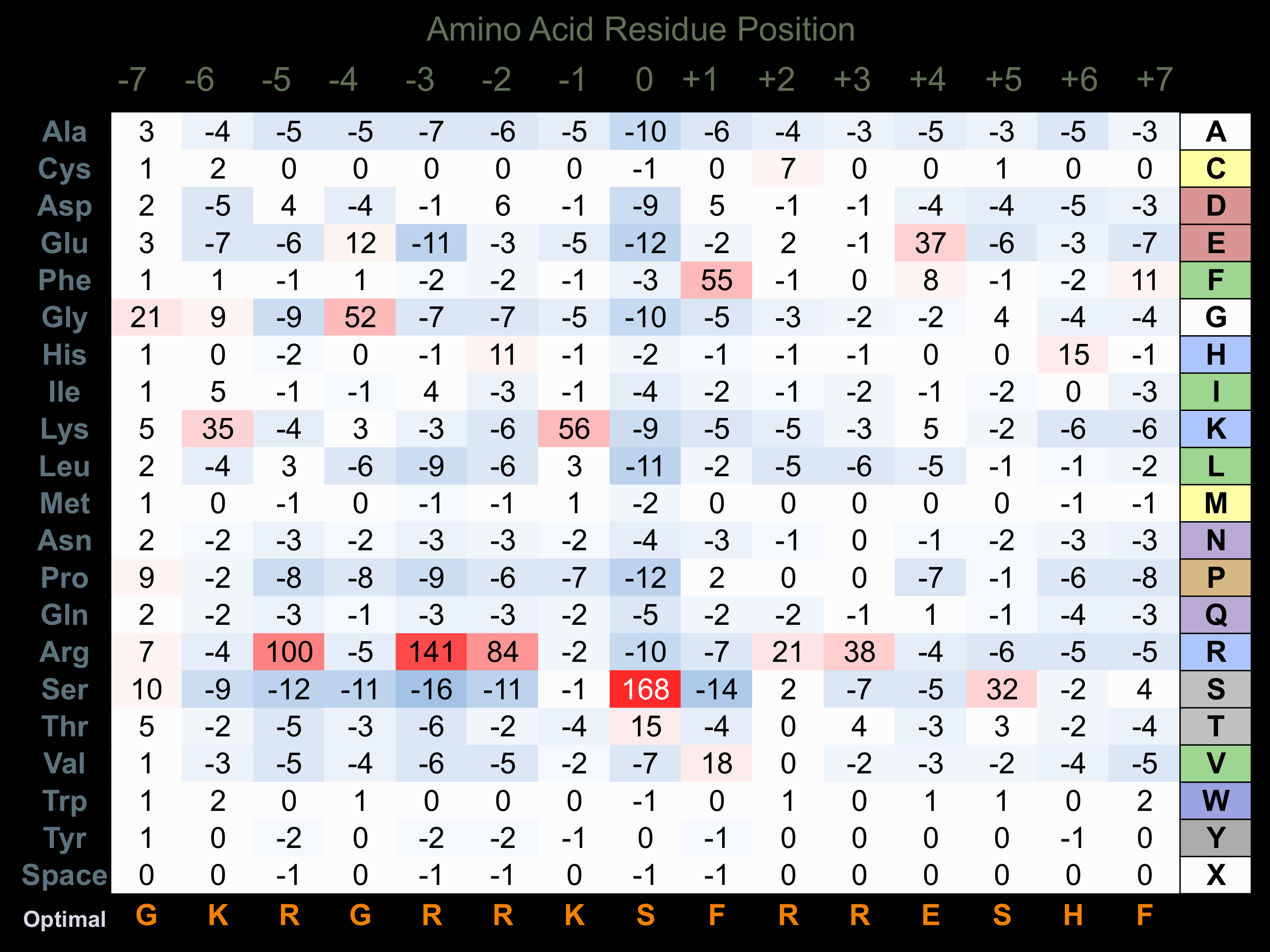
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Colorectal adenocarcinomas; Mantle cell lymphomas (LCM)
Comments:
Pim2 appears to be a oncoprotein (OP), although it shows a level of bystander mutations typical of most proteins in human cancers. The active form of the protein kinase normally acts to promote tumour cell proliferation. Pim2 is linked to Mantle Cell Lymphomas (LCM), which are non-Hodgkin lymphomas in which is specifically a B-cell lymphoma is also associated the ATM (ataxia telangiectasia mutated) gene.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +104, p<0.032); Brain oligodendrogliomas (%CFC= +48, p<0.007); Cervical cancer (%CFC= -56, p<0.003); Classical Hodgkin lymphomas (%CFC= +57, p<0.001); Gastric cancer (%CFC= -59, p<0.024); Large B-cell lymphomas (%CFC= +195, p<0.0007); Lung adenocarcinomas (%CFC= +68, p<0.001); and Oral squamous cell carcinomas (OSCC) (%CFC= +50, p<0.105).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 24433 diverse cancer specimens. This rate is the same as the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.4 % in 1052 large intestine cancers tested.
Frequency of Mutated Sites:
None > 1 in 20,654 cancer specimens
Comments:
Only 1 deletion and 1 insertion, and no complex mutations are noted on the COSMIC website. About 39% of the point mutations are silent and do not change the amino acid sequence of the protein kinase.

