Nomenclature
Short Name:
PITSLRE
Full Name:
PITSLRE serine-threonine-protein kinase CDC2L2
Alias:
- CD2L2
- CDK11-p58
- Cell division cycle 2-like 2
- EC 2.7.11.22
- Galactosyltransferase-associated protein kinase; P58GTA;
- CDC2L2
- CDC2L3
- CDK11A
- CDK11-p110
- CDK11-p46
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
CDK
SubFamily:
PITSLRE
Structure
Mol. Mass (Da):
90,974
# Amino Acids:
780
# mRNA Isoforms:
8
mRNA Isoforms:
91,362 Da (783 AA; Q9UQ88); 91,055 Da (779 AA; Q9UQ88-3); 91,018 Da (780 AA; Q9UQ88-2); 90,045 Da (770 AA; Q9UQ88-4); 62,316 Da (531 AA; Q9UQ88-9); 49,624 Da (439 AA; Q9UQ88-10); 45,229 Da (397 AA; Q9UQ88-5); 18,110 Da (167 AA; Q9UQ88-8)
4D Structure:
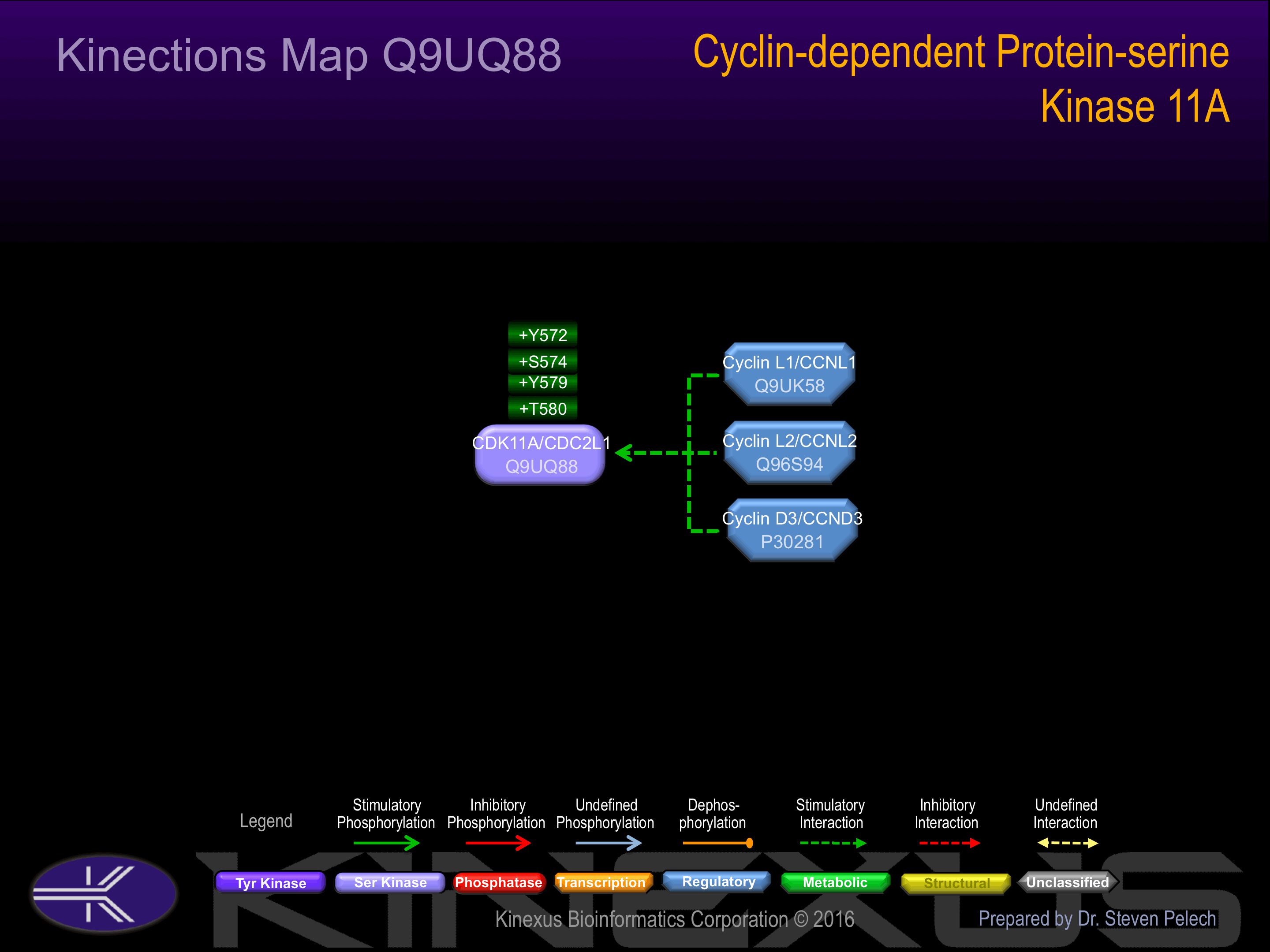
The cleaved p110 isoform, p110C, binds to the serine/threonine kinase PAK1. The p58 isoform but not the p110 isoform or p110C interacts with CCND3. The p110 isoforms are found in large molecular weight complexes containing CCNL1 and SFRS7.
1D Structure:
Subfamily Alignment
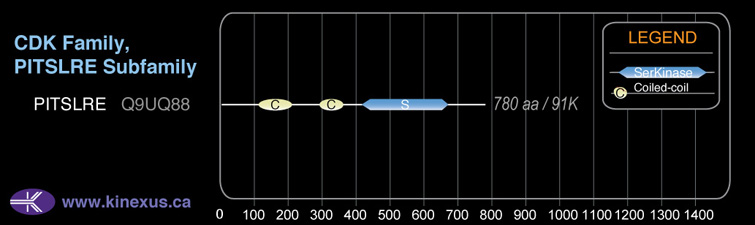
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 123 | 214 | Coiled-coil |
| 304 | 366 | Coiled-coil |
| 426 | 711 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K101, K123, K444, K446, K447, K455, .
Serine phosphorylated:
S47+, S65, S72, S92, S200, S217, S220, S222, S265, S271, S273, S369, S372, S398, S422, S574+, S577, S740, S751+, S773, S780.
Threonine phosphorylated:
T61, T436, T580+, T583, T588, T663, T739, T761, T766.
Tyrosine phosphorylated:
Y67, Y210, Y437, Y441+, Y572+, Y575+, Y579+, Y582, Y750+.
Ubiquitinated:
K20, K455, K565, K648, K707, K729.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
927
65
1001
 11
11
99
42
74
 10
10
92
5
84
 38
38
354
200
490
 93
93
863
60
690
 8
8
78
185
92
 41
41
376
85
562
 69
69
637
75
641
 70
70
652
44
518
 14
14
130
205
199
 13
13
116
60
110
 78
78
721
391
637
 10
10
93
60
80
 11
11
102
36
82
 10
10
96
45
95
 11
11
101
39
67
 14
14
131
403
61
 9
9
79
32
82
 12
12
114
203
102
 68
68
634
274
658
 15
15
142
40
126
 22
22
204
50
174
 8
8
73
10
87
 17
17
157
32
105
 25
25
234
40
282
 72
72
667
112
679
 9
9
84
69
63
 9
9
84
32
67
 13
13
119
32
76
 10
10
92
70
80
 16
16
147
30
116
 88
88
815
75
632
 19
19
175
85
112
 88
88
817
130
641
 30
30
281
65
462
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
0
0 0
0
0
0 -
-
-
- 97
97
-
- 98
98
-
- -
-
-
- -
-
-
- 97
97
-
- 91
91
-
- -
-
-
- -
-
-
- 98
98
-
- 85
85
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 40
40
-
- 49
49
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CCNL1 - Q9UK58 |
| 2 | PAK1 - Q13153 |
| 3 | RNPS1 - Q15287 |
| 4 | PAK1 - Q13153 |
| 5 | MYST2 - O95251 |
| 6 | EIF3F - O00303 |
| 7 | YWHAE - P62258 |
| 8 | YWHAG - P61981 |
| 9 | CASP1 - P29466 |
| 10 | CASP3 - P42574 |
| 11 | CARD17 - Q5XLA6 |
| 12 | CDK7 - P50613 |
| 13 | SF3A2 - Q15428 |
| 14 | POLR2A - P24928 |
Regulation
Activation:
phosphorylation at Thr-580 activates it
Inhibition:
Phosphorylation at Thr-433 or Tyr-434 inactivates the enzyme
Synthesis:
The p58 isoform is specifically induced in G2/M phase of the cell cycle.
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
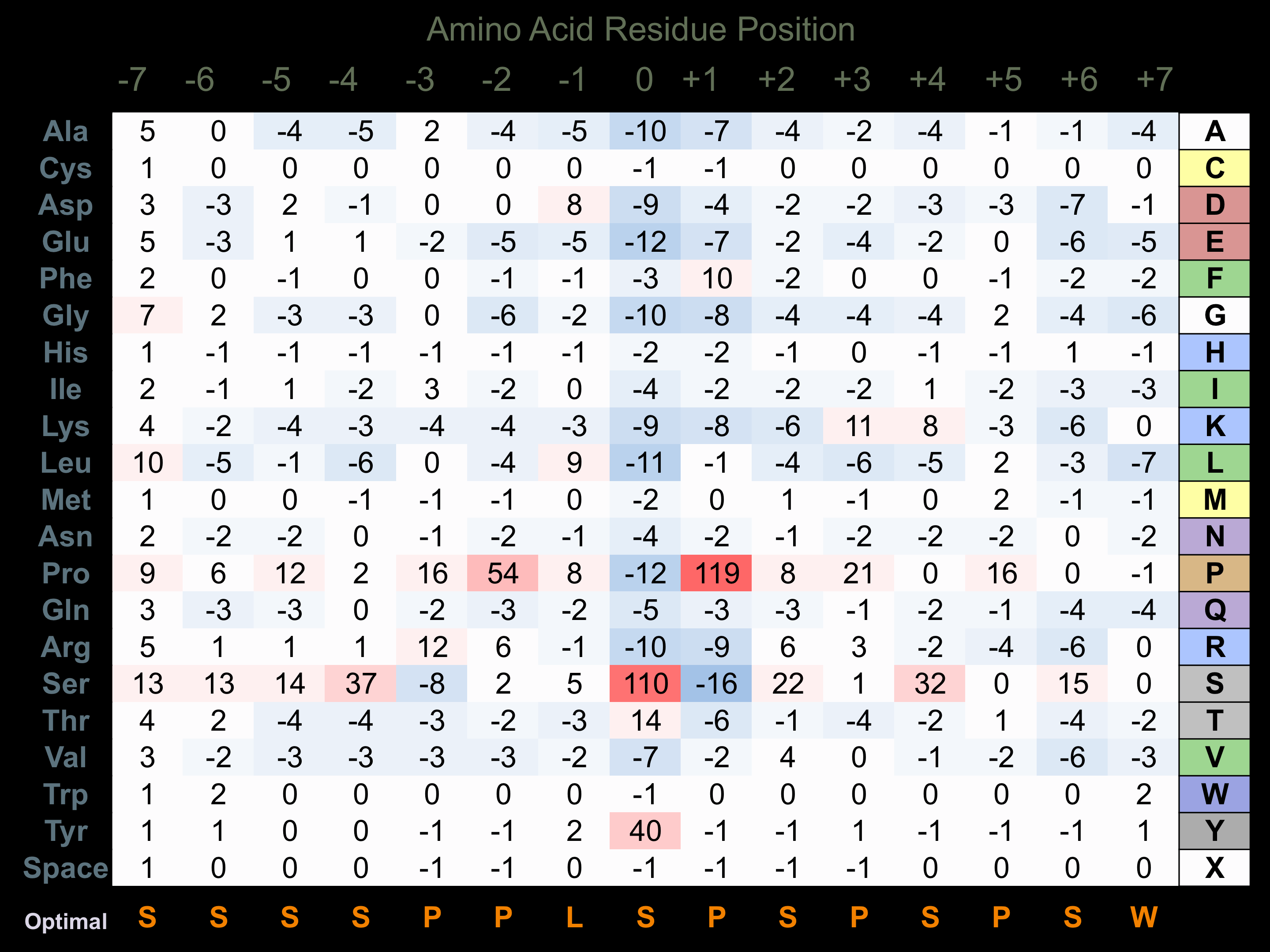
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| AT7519 | Kd = 5.2 nM | 11338033 | 22037378 | |
| AC1NS7CD | Kd = 48 nM | 5329665 | 295136 | 22037378 |
| SNS032 | Kd = 48 nM | 3025986 | 296468 | 18183025 |
| JNJ-7706621 | Kd = 110 nM | 5330790 | 191003 | 18183025 |
| Tozasertib | Kd = 260 nM | 5494449 | 572878 | 18183025 |
| Crizotinib | Kd = 420 nM | 11626560 | 601719 | 22037378 |
| Foretinib | Kd = 480 nM | 42642645 | 1230609 | 22037378 |
| Barasertib | Kd = 1 µM | 16007391 | 215152 | 18183025 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| AST-487 | Kd = 1.1 µM | 11409972 | 574738 | 18183025 |
| Linifanib | Kd = 1.2 µM | 11485656 | 223360 | 18183025 |
| Pazopanib | Kd = 1.3 µM | 10113978 | 477772 | 18183025 |
| R547 | Kd = 2.9 µM | 6918852 | 22037378 |
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Neuroblastomas (NB); Melanomas
Comments:
Advanced-stage neuroblastoma and tumour-derived cell contain a deletion of 1 allele of the gene. This gene was shown to be deleted or altered frequently in neuroblastoma. PITSLRE levels were also reported to be depressed in several cancer cell lines as well as in four malignant melanoma specimens.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +59, p<0.0002); Breast epithelial cell carcinomas (%CFC= +71, p<0.026); and Large B-cell lymphomas (%CFC= +59, p<0.097). The COSMIC website notes an up-regulated expression score for PITSLRE in diverse human cancers of 353, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 153 for this protein kinase in human cancers was 2.6-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.04 % in 24447 diverse cancer specimens. This rate is -52 % lower than the average rate of 0.075 % calculated for human protein kinases in general. Such a low frequency of mutation in human cancers is consistent with this protein kinase playing a role as a tumour requiring protein (TRP).
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.19 % in 603 endometrium cancers tested; 0.13 % in 1240 large intestine cancers tested; 0.1 % in 864 skin cancers tested; 0.09 % in 710 oesophagus cancers tested; 0.07 % in 548 urinary tract cancers tested; 0.07 % in 1300 breast cancers tested; 0.06 % in 1634 lung cancers tested; 0.05 % in 555 stomach cancers tested; 0.04 % in 1459 pancreas cancers tested; 0.03 % in 942 upper aerodigestive tract cancers tested; 0.03 % in 1512 liver cancers tested; 0.02 % in 558 thyroid cancers tested; 0.02 % in 1982 haematopoietic and lymphoid cancers tested; 0.01 % in 1253 kidney cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: D643E (8).
Comments:
Only 9 deletions (7 at E324_E326delEEE), and no insertions or no complex mutations are noted on the COSMIC website.