Nomenclature
Short Name:
PKACg
Full Name:
cAMP-dependent protein kinase, gamma-catalytic subunit
Alias:
- cAPKg
- PKAr
- PRKACG
- cAPKr
- KAPCG
- KAPG
- PKA C-gamma
- PKA-gamma
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
PKA
SubFamily:
NA
Structure
Mol. Mass (Da):
40434
# Amino Acids:
351
# mRNA Isoforms:
1
mRNA Isoforms:
40,434 Da (351 AA; P22612)
4D Structure:
A number of inactive tetrameric holoenzymes are produced by the combination of homo- or heterodimers of the different regulatory subunits associated with two catalytic subunits. cAMP causes the dissociation of the inactive holoenzyme into a dimer of regulatory subunits bound to four cAMP and two free monomeric catalytic subunits.
1D Structure:
Subfamily Alignment
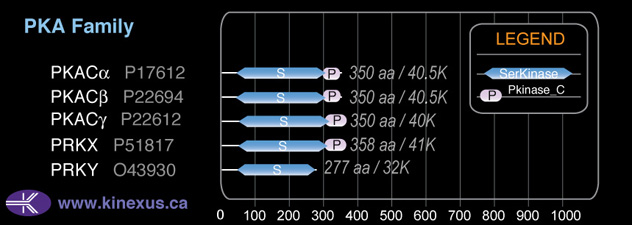
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
R132.
Myristoylated:
G2.
Threonine phosphorylated:
T196+, T198+, T202-.
Tyrosine phosphorylated:
Y118, Y123, Y205.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 19
19
1037
16
1386
 2
2
93
10
167
 0.4
0.4
24
1
0
 4
4
238
46
455
 8
8
403
14
406
 3
3
173
44
436
 4
4
205
19
389
 14
14
735
20
911
 5
5
256
10
259
 2
2
99
45
177
 2
2
93
14
184
 13
13
717
105
879
 0.3
0.3
18
12
16
 3
3
149
9
224
 0.8
0.8
41
10
64
 0.9
0.9
49
8
94
 0.9
0.9
50
99
46
 2
2
124
8
255
 0.4
0.4
20
43
16
 6
6
303
56
340
 1
1
53
10
62
 2
2
105
12
206
 8
8
404
10
259
 5
5
255
8
60
 3
3
142
10
273
 8
8
409
28
519
 0.3
0.3
15
15
9
 2
2
110
8
171
 1
1
55
8
72
 6
6
312
14
146
 7
7
389
18
276
 100
100
5368
21
8291
 3
3
173
46
462
 14
14
749
31
727
 7
7
375
22
276
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 76.6
76.6
76.8
99 54.7
54.7
57.3
- -
-
-
- -
-
-
100 78.9
78.9
88.6
- -
-
-
- 81.5
81.5
91.5
- 81.5
81.5
91.7
- -
-
-
- -
-
-
- 35.6
35.6
53.7
- 35.9
35.9
53.7
- 35.6
35.6
53.4
81.5 -
-
-
- 74.8
74.8
86.1
- -
-
-
- 61.4
61.4
73.8
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 44.2
44.2
64.7
49 -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PRKAR2A - P13861 |
| 2 | PRKAR2B - P31323 |
Regulation
Activation:
Activated by binding of two cAMP molecules to each of the two associated regulatory subunits in the PKA holoenzyme. Binding of cAMP induces dissociation of the two active catalytic subunits. Phosphorylation of Thr-198 increases phosphotransferase activity. Phosphorylation of S339 may play a role in stabilizing PKA.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
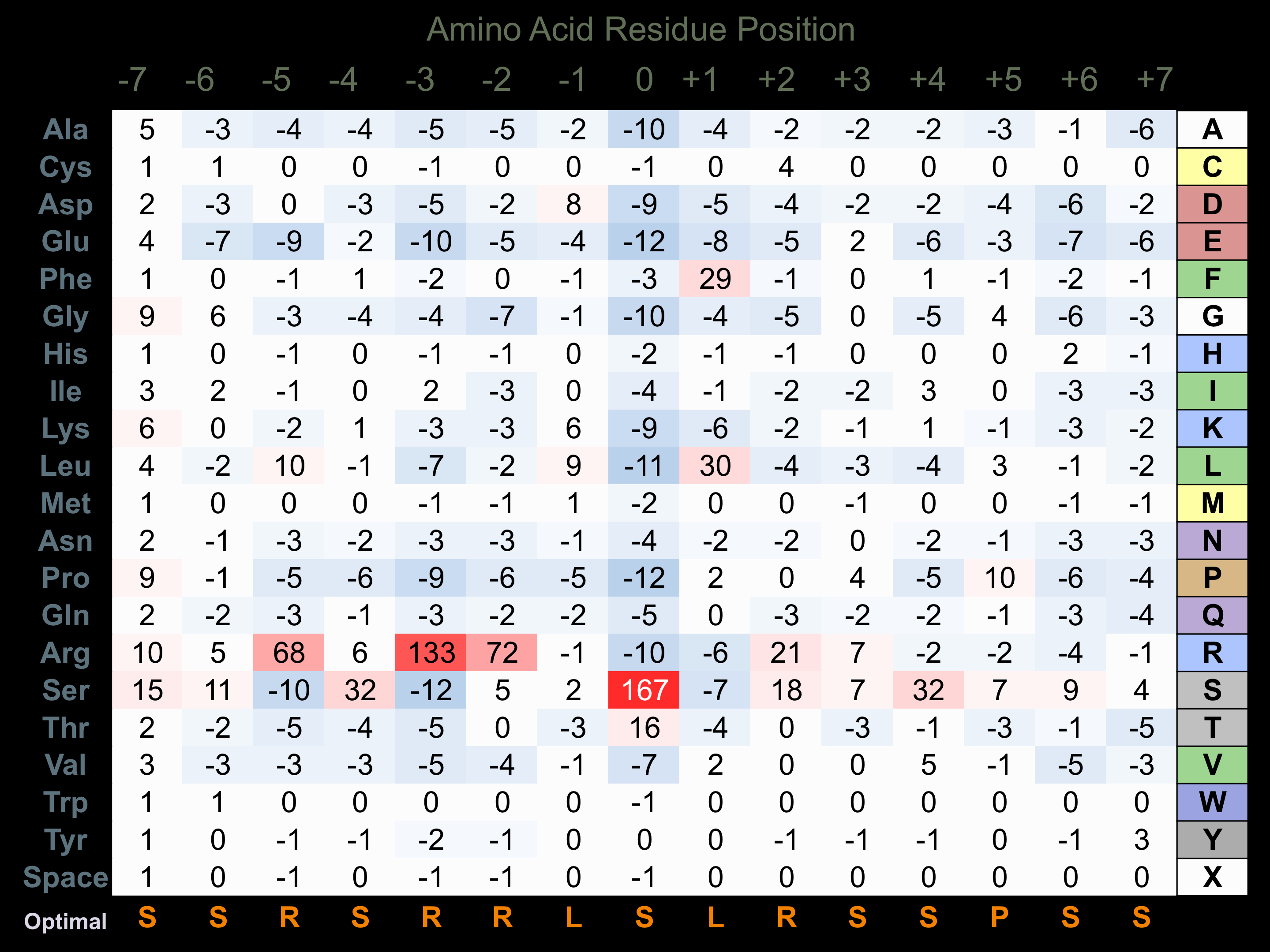
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| CHEMBL213618 | IC50 = 100 pM | 16043304 | 213618 | 16765046 |
| Staurosporine | IC50 = 900 pM | 5279 | 16765046 | |
| AC1OCAB6 | IC50 = 2.1 nM | 6914611 | 383264 | 16413780 |
| Ophiocordin | Ki = 4.7 nM | 5287736 | 60254 | 16134928 |
| Isoquinoline-pyridine; 10y | IC50 = 8 nM | 15604547 | 16603355 | |
| A674563 | Ki = 16 nM | 11314340 | 379218 | 16678413 |
| CHEMBL383541 | IC50 = 38 nM | 6914613 | 383541 | 16403626 |
| CHEMBL554986 | Kd = 48 nM | 11250806 | 554986 | 15634010 |
| K-252a; Nocardiopsis sp. | IC50 > 50 nM | 3813 | 281948 | 22037377 |
| CHEMBL534916 | Kd = 150 nM | 11490873 | 534916 | 15634010 |
| Meridianin B | IC50 = 210 nM | 10380356 | 15026054 | |
| SB218078 | IC50 > 250 nM | 447446 | 289422 | 22037377 |
| Balanol analog 5 | IC50 = 300 nM | 5327922 | 52529 | |
| Fasudil | Ki = 460 nM | 3547 | 38380 | 16249185 |
| PP1 Analog II; 1NM-PP1 | IC50 = 500 nM | 5154691 | 573578 | 22037377 |
| CK7 | Ki > 1 µM | 447961 | 15027857 | |
| H-89 | IC50 = 1.2 µM | 449241 | 104264 | 16890431 |
| CHEMBL536485 | IC50 = 2.4 µM | 11445419 | 536485 | 15634010 |
| N-Benzoylstaurosporine | IC50 = 2.4 µM | 56603681 | 608533 | |
| Purvalanol B | IC50 = 3.8 µM | 448991 | 23254 | 12036347 |
Disease Linkage
General Disease Association:
Endocrine disorders
Specific Diseases (Non-cancerous):
Primary pigmented nodular adrenocortical disease (PPNAD); Cushing syndrome, ACTH-independent adrenal, somatic
Comments:
There is a possible association between duplication of the PRKACG gene and 46,XY gonadal dysgenesis.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= -46, p<0.011); Breast epithelial carcinomas (%CFC= -59, p<0.059); and Uterine leiomyomas (%CFC= +120, p<0.005). The COSMIC website notes an up-regulated expression score for PKACg in diverse human cancers of 422, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.15 % in 25431 diverse cancer specimens. This rate is 2-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 1.06 % in 864 skin cancers tested; 0.61 % in 603 endometrium cancers tested; 0.56 % in 1270 large intestine cancers tested; 0.34 % in 589 stomach cancers tested; 0.31 % in 548 urinary tract cancers tested; 0.2 % in 710 oesophagus cancers tested; 0.19 % in 441 autonomic ganglia cancers tested; 0.11 % in 1276 kidney cancers tested; 0.1 % in 833 ovary cancers tested; 0.1 % in 273 cervix cancers tested; 0.1 % in 1490 breast cancers tested; 0.09 % in 942 upper aerodigestive tract cancers tested; 0.09 % in 2009 haematopoietic and lymphoid cancers tested; 0.09 % in 1512 liver cancers tested; 0.07 % in 1957 lung cancers tested; 0.06 % in 939 prostate cancers tested; 0.06 % in 1467 pancreas cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,714 cancer specimens
Comments:
Only 2 deletions, no insertions, and 3 complex mutations are noted on the COSMIC website.

