Nomenclature
Short Name:
PKG1
Full Name:
cGMP-dependent protein kinase 1, alpha isozyme
Alias:
- cGK 1 alpha
- EC 2.7.11.12
- KGP1A
- Kinase PKG1
- MGC71944
- PGK; PKG; PKG1-alpha; Protein kinase, cGMP-dependent, type I
- cGKI-alpha
- CGKI-beta
- cGMP-dependent protein kinase 1, alpha isozyme
- DKFZp686K042
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
PKG
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
76364
# Amino Acids:
671
# mRNA Isoforms:
3
mRNA Isoforms:
77,804 Da (686 AA; Q13976-2); 76,364 Da (671 AA; Q13976); 44,185 Da (389 AA; Q13976-3)
4D Structure:
Isoform alpha: parallel homodimer or heterodimer and also heterotetramer. Interacts directly with PPP1R12A. Non-covalent dimer of dimer of PRKG1-PRKG1 and PPP1R12A-PPP1R12A. This interaction targets PRKG1 to stress fibers to mediate smooth muscle cell relaxation and vasodilation in responses to rises in cGMP. Isoform beta: antiparallel homodimer. Part of cGMP kinase signaling complex at least composed of ACTA2/alpha-actin, CNN1/calponin H1, PLN/phospholamban, PRKG1 and ITPR1 By similarity. Interacts with MRVI1
1D Structure:
Subfamily Alignment
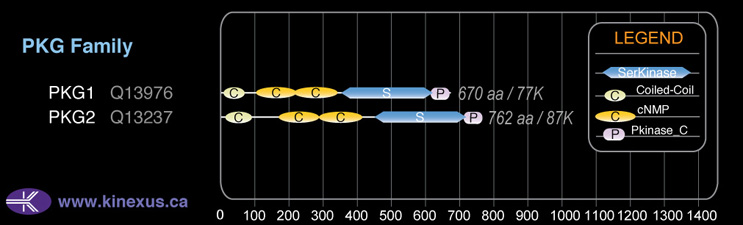
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 10 | 62 | Coiled-coil |
| 102 | 219 | cNMP |
| 220 | 340 | cNMP |
| 360 | 619 | Pkinase |
| 620 | 671 | Pkinase_C |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
S2, K607, K615, K617.
Serine phosphorylated:
S27, S45, S51, S65+, S73, S218, S231, S273.
Threonine phosphorylated:
T59+, T85, T515+, T517+, T521-.
Tyrosine phosphorylated:
Y212, Y247, Y433, Y435, Y497.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 34
34
1067
38
1281
 1
1
31
18
45
 4
4
122
11
144
 5
5
167
116
312
 15
15
476
36
471
 2
2
63
90
186
 4
4
117
44
280
 12
12
362
43
474
 6
6
199
20
196
 2
2
69
108
84
 1
1
31
34
66
 17
17
516
194
583
 0.8
0.8
26
33
60
 0.8
0.8
24
15
21
 1
1
30
18
45
 2
2
51
20
70
 4
4
121
188
1037
 1.3
1.3
42
23
84
 6
6
200
107
281
 13
13
397
140
460
 2
2
51
27
88
 1.1
1.1
35
29
92
 1.5
1.5
46
21
63
 1.2
1.2
36
23
65
 1
1
32
27
83
 16
16
497
74
615
 0.7
0.7
21
39
50
 1.3
1.3
41
21
107
 3
3
97
22
133
 3
3
84
42
97
 12
12
386
24
394
 100
100
3126
53
6078
 3
3
91
75
324
 19
19
603
83
575
 4
4
128
48
171
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
100 -
-
-
100 -
-
-
93 -
-
-
- 68.6
68.6
72.2
93 -
-
-
- 99.5
99.5
99.8
93 48.6
48.6
65.2
93 -
-
-
- -
-
-
- -
-
-
93 -
-
-
96 -
-
-
85 -
-
-
- 40.9
40.9
51.1
63 61.2
61.2
75.9
- 45.1
45.1
61.6
49 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 25.4
25.4
39.7
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | MRVI1 - Q9Y6F6 |
| 2 | PPP1R12A - O14974 |
| 3 | VASP - P50552 |
| 4 | FHOD1 - Q9Y613 |
| 5 | PLCB3 - Q01970 |
| 6 | GTF2I - P78347 |
| 7 | RGS2 - P41220 |
| 8 | RAF1 - P04049 |
| 9 | TNNT1 - P13805 |
| 10 | PRKCA - P17252 |
| 11 | GAPDH - P04406 |
| 12 | CREB1 - P16220 |
| 13 | SPP1 - P10451 |
| 14 | CFTR - P13569 |
Regulation
Activation:
Binding of cGMP stimulates the phosphotransferase activity of PKG1. Phosphorylation at Thr-59 increases phosphotransferase activity and affinity for cGMP Phosphorylation at Thr-517 increases phosphotransferase activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| AKAP12 | Q02952 | S627 | KKRVRRPSESDKEDE | |
| Arp3 | P61158 | T257 | SKWIKQYTGINAISK | |
| CFTR | P13569 | S660 | FSAERRNSILTETLH | |
| CFTR | P13569 | S700 | FGEKRKNSILNPINS | |
| CRIP2 | P52943 | S104 | RAEERKASGPPKGPS | |
| FHOD1 | Q9Y613 | S1130 | AARERKRSRGNRKSL | |
| GABRB1 | P18505 | S434 | GRIRRRASQLKVKIP | |
| GCAP2 | Q9UMX6 | S197 | LAQQRRKSAMF____ | |
| GSBS | O96001 | T119 | KKPRRKDTPALHMSP | |
| GSBS | O96001 | T72 | KKPRRKDTPALHIPP | |
| GSK3b | P49841 | S9 | SGRPRTTSFAESCKP | - |
| GTF2I | P78347 | S412 | GIPFRRPSTYGIPRL | |
| GTF2I | P78347 | S784 | GVPFRRPSTFGIPRL | |
| GUCY1A3 | Q02108 | S65 | SLPQRKTSRSRVYLH | - |
| H1D | P16403 | S36 | GGTPRKASGPPVSEL | |
| H1R | P35367 | S396 | FTWKRLRSHSRQYVS | |
| H1R | P35367 | S398 | WKRLRSHSRQYVSGL | |
| H2B | P33778 | S33 | DGKKRKRSRKESYSI | |
| H2B | P33778 | S37 | RKRSRKESYSIYVYK | |
| HSL | Q05469 | S853 | IAEPMRRSVSEAALA | |
| HSP20 | O14558 | S16 | PSWLRRASAPLPGLS | ? |
| HSP27 | P04792 | S15 | FSLLRGPSWDPFRDW | ? |
| HSP27 | P04792 | S78 | PAYSRALSRQLSSGV | + |
| HSP27 | P04792 | S82 | RALSRQLSSGVSEIR | ? |
| HSP27 | P04792 | T143 | RCFTRKYTLPPGVDP | |
| IP3R1 (IP3 receptor) | Q14643 | S1598 | RNAARRDSVLAASRD | |
| IP3R1 (IP3 receptor) | Q14643 | S1764 | RPSGRRESLTSFGNG | |
| KCNMA1 | Q12791 | S1204 | QSSSKKSSSVHSIPS | |
| Lasp-1 | Q14847 | S146 | MEPERRDSQDGSSYR | |
| MDH1 | P40925 | S240 | VIKARKLSSAMSAAK | |
| mGluR7 | Q14831 | S862 | NVQKRKRSFKAVVTA | |
| MYLK1 (smMLCK) | Q15746 | S1005 | GLSGRKSSTGSPTSP | |
| MYLK1 (smMLCK) | Q15746 | S1773 | GLSGRKSSTGSPTSP | |
| MYLK1 (smMLCK) | Q15746 | S1779 | SSTGSPTSPLNAEKL | |
| MYPT1 | O14974 | S695 | QARQSRRSTQGVTLT | |
| MYPT1 | O14974 | S852 | RPREKRRSTGVSFWT | |
| MYPT1 | O14974 | T696 | ARQSRRSTQGVTLTD | |
| PAK1 | Q13153 | S21 | APPMRNTSTMIGAGS | - |
| PDE5A | O76074 | S102 | GTPTRKISASEFDRP | + |
| PKAR1A | P10644 | S101 | RRRRGAISAEVYTEE | |
| PKG1 (PRKG1) | Q13976 | S27 | KELEKRLSEKEEEIQ | |
| PKG1 (PRKG1) | Q13976 | S45 | RKLHKCQSVLPVPST | |
| PKG1 (PRKG1) | Q13976 | S51 | QSVLPVPSTHIGPRT | |
| PKG1 (PRKG1) | Q13976 | S65 | TTRAQGISAEPQTYR | + |
| PKG1 (PRKG1) | Q13976 | S73 | AEPQTYRSFHDLRQA | |
| PKG1 (PRKG1) | Q13976 | T517 | GFGKKTWTFCGTPEY | + |
| PKG1 (PRKG1) | Q13976 | T59 | THIGPRTTRAQGISA | + |
| PKG1 (PRKG1) | Q13976 | T85 | RQAFRKFTKSERSKD | |
| PLCB3 | Q01970 | S1105 | LDRKRHNSISEAKMR | - |
| PLCB3 | Q01970 | S26 | VETLRRGSKFIKWDE | - |
| PLCB3 | Q01970 | T1122 | HKKEAELTEINRRHI | - |
| PTRF | Q6NZI2 | S300 | SRDKLRKSFTPDHVV | |
| PTRF | Q6NZI2 | S365 | AGDLRRGSSPDVHAL | |
| PTRF | Q6NZI2 | S366 | GDLRRGSSPDVHALL | |
| PTRF | Q6NZI2 | T302 | DKLRKSFTPDHVVYA | |
| PTS | Q03393 | S19 | AQVSRRISFSASHRL | |
| Rap1b | P61224 | S179 | PGKARKKSSCQLL__ | |
| Rap1GAP2 | Q684P5 | S7 | _MFGRKRSVSFGGFG | |
| RGS4 | P49798 | S52 | VVICQRVSQEEVKKW | |
| RhoA | P61586 | S188 | ARRGKKKSGCLVL__ | |
| RIL | P50479 | S120 | PTTSRRPSGTGTGPE | |
| RyR1 | P21817 | S2843 | KKKTRKISQSAQTYD | |
| RyR2 | Q92736 | S2806 | YNRTRRISQTSQVSV | |
| Septin-3 | Q9UH03 | S91 | SQVSRKASSWNREEK | |
| SERPINA1 | P01009 | T51 | HPTFNKITPNLAEFA | |
| SERT | P31645 | T276 | SIWKGVKTSGKVVWV | |
| TBXA2R iso2 | P21731-2 | S330 | LSTRPRRSLTLWPSL | |
| TBXA2R iso2 | P21731-2 | T332 | TRPRRSLTLWPSLEY | |
| Titin | Q8WZ42 | S4185 | IQQGAKTSLQEEMDS | |
| Titin | Q8WZ42 | S4372 | DSGKTATSAKLTVVK | |
| TNNI3 | P19429 | S22 | PAPIRRRSSNYRAYA | |
| TNNI3 | P19429 | S23 | APIRRRSSNYRAYAT | |
| TRPC3 | Q13507 | S263 | KNDYRKLSMQCKDFV | |
| TRPC3 | Q13507 | T11 | SPSLRRMTVMREKGR | |
| TRPC6 | Q9Y210 | T70 | RLAHRRQTVLREKGR | - |
| VASP | P50552 | S156 | EHIERRVSNAGGPPA | |
| VASP | P50552 | S238 | GAKLRKVSKQEEASG | |
| VASP | P50552 | T277 | LARRRKATQVGEKTP |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
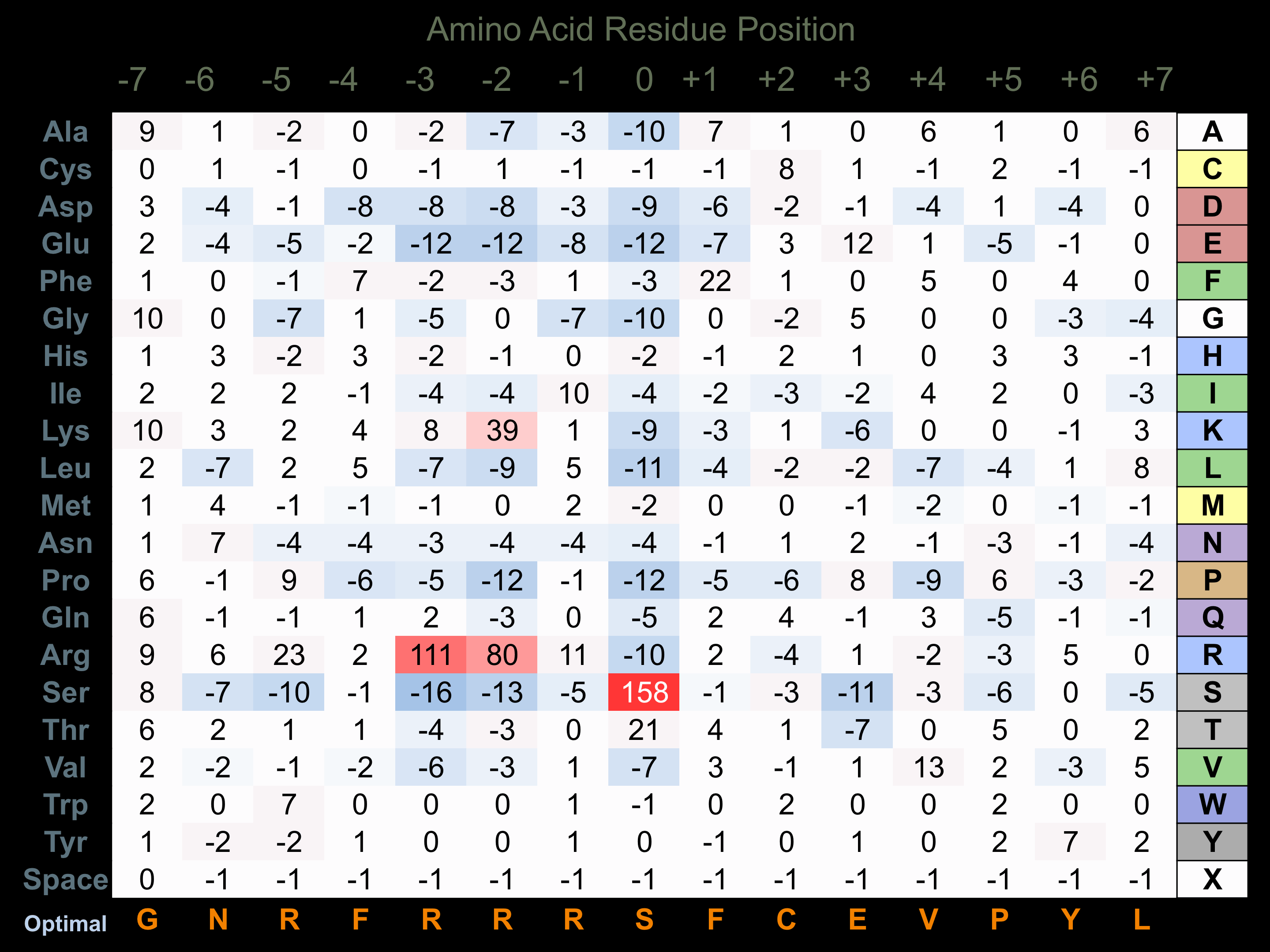
Matrix Type:
Experimentally derived from alignment of 116 known protein substrate phosphosites and 12 peptides phosphorylated by recombinant PKG1 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, cardiovascular disorders
Specific Diseases (Non-cancerous):
Aortic aneurysm, familial thoracic 8; Erdheim-Chester disease; Phosphoglycerate kinase deficiency; Aortic aneurysm, familial thoracic 4
Comments:
Aortic aneurysm, familial thoracic 8 (AAT8) is characterized by permanent dilation of the thoracic aorta. This is usually due to degeneration of and fragmentation of elastic fibers, loss of smooth muscle cells, and an accumulation of basophilic ground substance in of the aortic wall, which features a characteristic histologic appearance known as 'medial necrosis' or 'Erdheim cystic medial necrosis.'
Specific Cancer Types:
Hemangiomas; Intraductal papillomas
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= -52, p<0.036); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +1207, p<0.028); Colon mucosal cell adenomas (%CFC= -48, p<0.0002); Ovary adenocarcinomas (%CFC= -54, p<0.032); and Prostate cancer - metastatic (%CFC= -59, p<0.0001). The COSMIC website notes an up-regulated expression score for PKG1 in diverse human cancers of 369, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. An R177Q mutation in AAT8 results in a constitutively active form of PKG1, which also has reduced cGMP binding.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.12 % in 24934 diverse cancer specimens. This rate is 1.63-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.74 % in 864 skin cancers tested; 0.4 % in 1270 large intestine cancers tested; 0.33 % in 1634 lung cancers tested; 0.3 % in 589 stomach cancers tested; 0.25 % in 603 endometrium cancers tested; 0.18 % in 1512 liver cancers tested; 0.14 % in 548 urinary tract cancers tested; 0.12 % in 127 biliary tract cancers tested; 0.09 % in 942 upper aerodigestive tract cancers tested; 0.08 % in 710 oesophagus cancers tested; 0.08 % in 1316 breast cancers tested; 0.07 % in 833 ovary cancers tested; 0.06 % in 238 bone cancers tested; 0.05 % in 558 thyroid cancers tested; 0.05 % in 273 cervix cancers tested; 0.04 % in 2009 haematopoietic and lymphoid cancers tested; 0.04 % in 1276 kidney cancers tested; 0.03 % in 881 prostate cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: A354V (3); S470L (3).
Comments:
Only 1 deletion, no insertion, and 1 complex mutation are noted on the COSMIC website.

