Nomenclature
Short Name:
PRKX
Full Name:
Serine-threonine-protein kinase PRKX
Alias:
- EC 2.7.11.1
- PKX1
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
PKA
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
40,896
# Amino Acids:
358
# mRNA Isoforms:
1
mRNA Isoforms:
40,896 Da (358 AA; P51817)
4D Structure:
NA
1D Structure:
Subfamily Alignment
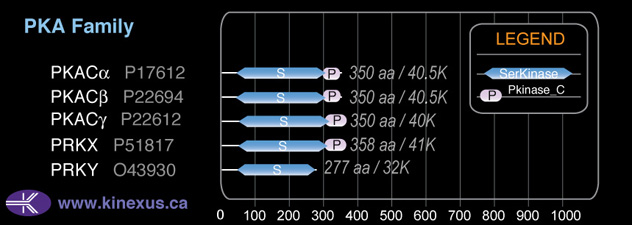
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S305.
Threonine phosphorylated:
T201+, T203+, T207.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 14
14
750
16
961
 0.2
0.2
11
8
5
 1
1
54
9
53
 26
26
1392
60
3329
 11
11
600
14
587
 5
5
262
44
516
 6
6
300
19
482
 27
27
1482
31
2935
 4
4
243
10
203
 1.3
1.3
69
50
45
 0.7
0.7
38
20
44
 10
10
559
101
587
 2
2
113
20
127
 1.2
1.2
63
6
20
 0.6
0.6
34
9
17
 0.5
0.5
25
7
16
 0.6
0.6
32
104
35
 2
2
123
14
231
 0.5
0.5
29
45
19
 9
9
498
56
534
 2
2
101
16
71
 2
2
108
18
89
 1.1
1.1
59
18
75
 1.1
1.1
62
14
41
 2
2
101
16
94
 21
21
1157
38
1926
 1.3
1.3
71
23
59
 2
2
109
14
100
 1.1
1.1
62
14
40
 1.1
1.1
58
14
56
 20
20
1083
18
689
 100
100
5444
21
8322
 14
14
748
53
1111
 15
15
800
31
808
 2
2
111
22
56
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 93
93
95.5
93 98
98
98.3
98 -
-
-
86 -
-
-
- 80.2
80.2
85.5
91.5 -
-
-
- 77.4
77.4
84.9
78 49.2
49.2
68.4
81 -
-
-
- 66
66
72
- 81.4
81.4
88
89.5 31.7
31.7
46.3
81 73.7
73.7
84.4
76.5 -
-
-
- 38.4
38.4
49.9
62.5 56.7
56.7
72.2
- 42.1
42.1
61.1
51 47.8
47.8
58.7
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 43.1
43.1
63
48 -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PRKAR2A - P13861 |
| 2 | PRKAR1A - P10644 |
| 3 | PKIA - P61925 |
| 4 | PRKAR1B - P31321 |
| 5 | PRKAR2B - P31323 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
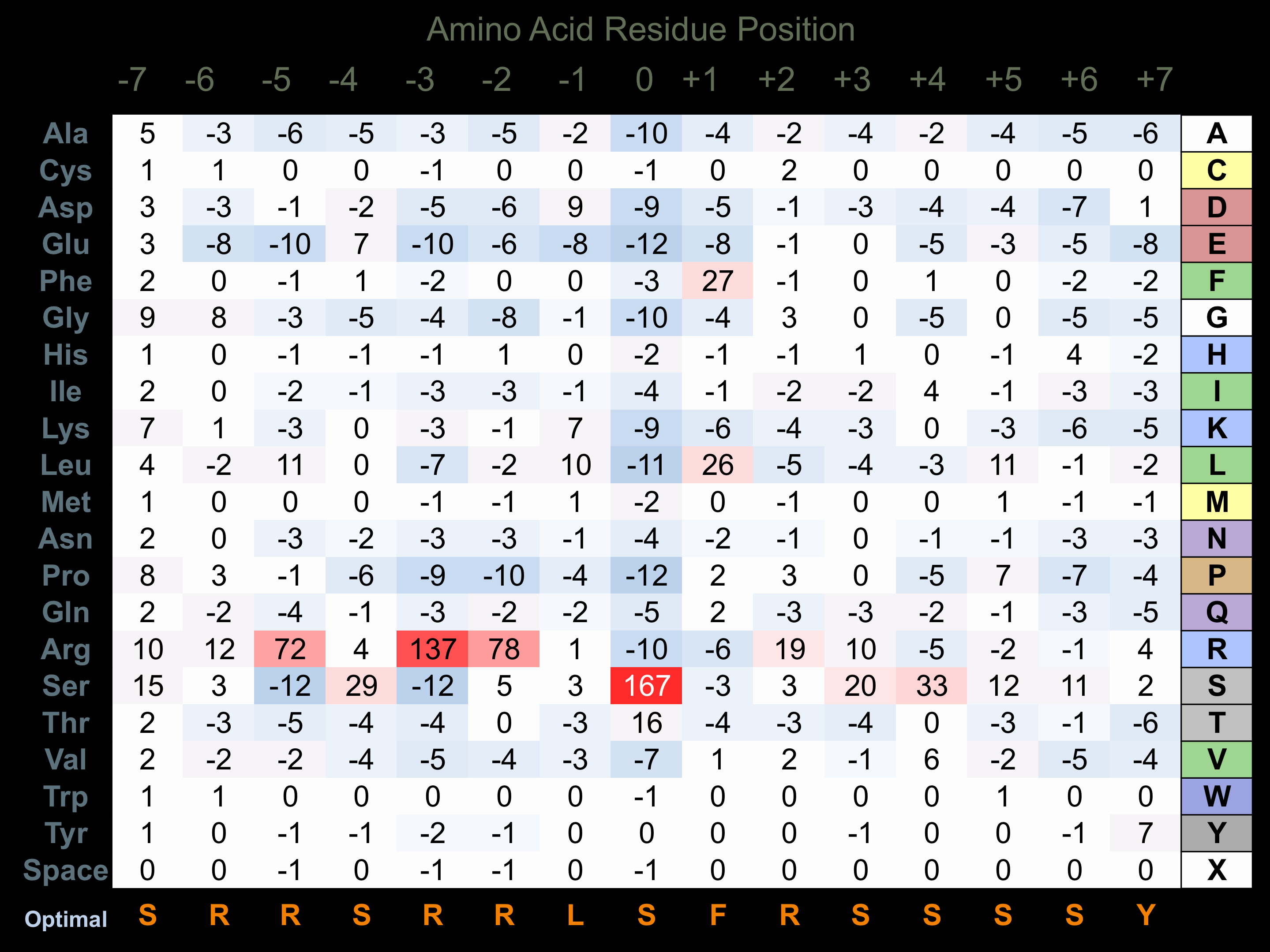
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Nephrological, and reproductive disorders
Specific Diseases (Non-cancerous):
Polycystic kidney disease (PKD1, PKD); Sex reversal disorder
Comments:
Polycystic Kidney Disease (PKD1, PKD) is characterized by formation of cysts in the kidney, which can possibly lead to kidney failure. An estimated 30% of Sex Reversal Disorder in XY females and XX males are caused by a chromosomal translocation across the PRKX gene. The K78R mutation leads to impaired PRKX function. The H93Q and W202R, in conjunction lead to constitutive serine/threonine kinase phosphotransferase activity. R283L is a mutation that will increase binding of PRKX to PRKAR2A and PRKAR2B.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= -74, p<0.012); Brain oligodendrogliomas (%CFC= -89, p<0.004); Ovary adenocarcinomas (%CFC= +93, p<0.024); Pituitary adenomas (ACTH-secreting) (%CFC= -61); and Prostate cancer (%CFC= +64, p<0.007). The COSMIC website notes an up-regulated expression score for PRKX in diverse human cancers of 407, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 149 for this protein kinase in human cancers was 2.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24914 diverse cancer specimens. This rate is only -4 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.36 % in 864 skin cancers tested; 0.24 % in 589 stomach cancers tested; 0.23 % in 603 endometrium cancers tested; 0.2 % in 548 urinary tract cancers tested; 0.2 % in 1270 large intestine cancers tested; 0.12 % in 1822 lung cancers tested; 0.11 % in 1316 breast cancers tested; 0.08 % in 710 oesophagus cancers tested; 0.07 % in 1512 liver cancers tested; 0.06 % in 1459 pancreas cancers tested; 0.04 % in 1276 kidney cancers tested; 0.03 % in 2082 central nervous system cancers tested.
Frequency of Mutated Sites:
None > 6 in 20,197 cancer specimens
Comments:
No deletions, insertions or complex mutations are noted on the COSMIC website.

