Nomenclature
Short Name:
PRP4
Full Name:
Serine-threonine-protein kinase PRP4 homolog
Alias:
- CBP143
- Pre-mRNA protein kinase
- PRPF4B
- PRP4 homolog
- PRP4 kinase
- PRPK
- EC 2.7.11.1
- KIAA0536
- Kinase PRP4
- PR4H
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
DYRK
SubFamily:
PRP4
Specific Links
Structure
Mol. Mass (Da):
116,973
# Amino Acids:
1007
# mRNA Isoforms:
1
mRNA Isoforms:
116,987 Da (1007 AA; Q13523)
4D Structure:
Identified in the spliceosome C complex, at least composed of AQR, ASCC3L1, C19orf29, CDC40, CDC5L, CRNKL1, DDX23, DDX41, DDX48, DDX5, DGCR14, DHX35, DHX38, DHX8, EFTUD2, FRG1, GPATC1, HNRPA1, HNRPA2B1, HNRPA3, HNRPC, HNRPF, HNRPH1, HNRPK, HNRPM, HNRPR, HNRPU, KIAA1160, KIAA1604, LSM2, LSM3, MAGOH, MORG1, PABPC1, PLRG1, PNN, PPIE, PPIL1, PPIL3, PPWD1, PRPF19, PRPF4B, PRPF6, PRPF8, RALY, RBM22, RBM8A, RBMX, SART1, SF3A1, SF3A2, SF3A3, SF3B1, SF3B2, SF3B3, SFRS1, SKIV2L2, SNRPA1, SNRPB, SNRPB2, SNRPD1, SNRPD2, SNRPD3, SNRPE, SNRPF, SNRPG, SNW1, SRRM1, SRRM2, SYF2, SYNCRIP, TFIP11, THOC4, U2AF1, WDR57, XAB2 and ZCCHC8. Interacts with Clk1 C-terminus.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
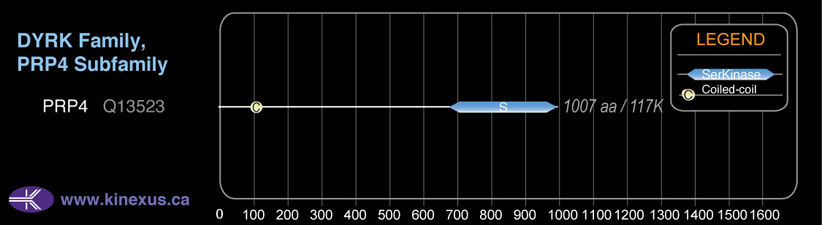
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 101 | 122 | Coiled-coil |
| 687 | 1003 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K168, K177, K200, K202, K358 (N6), K656, K717 (N6), K905, K907.
Serine phosphorylated:
S8, S20, S23, S32, S36, S41, S87, S93, S131, S142, S144, S164, S165, S166, S184, S196, S201, S239, S241, S257, S277, S283, S285, S292, S294, S328, S330, S335, S349, S354, S356, S366, S368, S381, S383, S387, S394, S396, S410, S411, S427, S431, S437, S443, S445, S458, S460, S507, S509, S518, S519, S520, S562, S565, S568, S569, S572, S578, S580, S606, S622, S623, S636, S794, S837+, S839+, S852-.
Threonine phosphorylated:
T162, T243, T259, T385, T414, T439, T574, T576, T847+.
Tyrosine phosphorylated:
Y140, Y552, Y554, Y643, Y674, Y849+, Y855-.
Ubiquitinated:
K182.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 35
35
1027
48
1135
 2
2
47
23
40
 9
9
263
14
228
 19
19
560
160
1005
 30
30
870
45
709
 4
4
107
126
130
 9
9
277
57
419
 34
34
995
60
2167
 27
27
804
24
576
 3
3
100
152
133
 5
5
152
42
204
 24
24
711
263
680
 8
8
223
47
269
 2
2
63
18
55
 3
3
83
25
62
 4
4
110
28
87
 5
5
153
223
1363
 6
6
168
27
227
 5
5
134
146
117
 24
24
702
190
706
 5
5
143
32
163
 9
9
253
38
277
 7
7
220
25
251
 5
5
144
27
164
 9
9
261
33
316
 27
27
797
99
699
 7
7
202
50
198
 5
5
159
27
186
 8
8
240
27
284
 1.4
1.4
41
56
41
 27
27
795
30
398
 100
100
2944
56
6785
 17
17
508
101
730
 33
33
963
109
782
 8
8
236
61
353
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 92.6
92.6
92.6
100 99
99
99.3
99 -
-
-
97 -
-
-
96 95.3
95.3
96.2
98 -
-
-
- 95.1
95.1
97.5
95 95.4
95.4
97.8
95 -
-
-
- 93.1
93.1
96.5
- 89.8
89.8
94
90 80.6
80.6
89.5
80 68.8
68.8
78.4
- -
-
-
- 41.4
41.4
57.4
43 -
-
-
- 36.8
36.8
51.9
41 48.4
48.4
60
- -
-
-
- -
-
-
- -
-
-
48 -
-
-
33 -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| AHNAK | Q09666 | S177 | AKDIDISSPEFKIKI | |
| ATRIP | Q8WXE1 | S224 | APSVSHVSPRKNPSV | |
| BAIAP2L1 (IRTKS) | Q9UHR4 | S261 | VSGTPQASPMIERSN | |
| BCLAF1 | Q9NYF8 | S512 | LKDLFDYSPPLHKNL | |
| CTTN (Cortactin) | Q14247 | S405 | KTQTPPVSPAPQPTE | - |
| DPF2 | Q92785 | S142 | DPRVDDDSLGEFPVT | |
| EEF1D | P29692 | S162 | DDIDLFGSDNEEEDK | |
| Elk-1 | P19419 | T417 | ISVDGLSTPVVLSPG | |
| G3BP1 | Q13283 | S232 | EDAQKSSSPAPADIA | - |
| GATAD2B | Q8WXI9 | S122 | ERGRLTPSPDIIVLS | |
| hnRNP K | P61978 | S216 | ILDLISESPIKGRAQ | + |
| IGF2BP2 | Q9Y6M1 | S164 | DEEVSSPSPPQRAQR | |
| MCM2 | P49736 | S27 | GNDPLTSSPGRSSRR | ? |
| NCOR2 (SMRT) | Q9Y618 | S956 | GDPRANASPQkPLDL | |
| PAK4 | O96013 | S104 | SNSLRRDSPPPPARA | |
| PRKAR1A | P10644 | S83 | DSREDEISPPPPNPV | - |
| PSMD2 | Q13200 | S16 | APVQPQQSPAAAPGG | |
| RBM17 (SF45) | Q96I25 | S155 | ARRPDPDSDEDEDYE | |
| RNF20 | Q5VTR2 | S138 | EPEPDSDSNQERKDD | |
| RPL12 | P30050 | S38 | KIGPLGLSPKKVGDD | |
| SF1 | Q15637 | S80 | PPNPEDRSPSPEPIY | |
| SF1 | Q15637 | S82 | NPEDRSPSPEPIYNS | |
| SH3KBP1 | Q96B97 | S587 | TAGHRANSPSLFGTE | - |
| SNRNP70 | P08621 | S226 | YDERPGPSPLPHRDR | |
| SRRM2 (SRm300) | Q9UQ35 | S377 | LAERHGGSPQPLATT | |
| SRSF6 | Q13247 | S303 | RSQSRSNSPLPVPPS | |
| TBC1D10B | Q4KMP7 | S678 | RAAGGAPSPPPPVRR | |
| TCOF1 (Treacle) | Q13428 | S1378 | EGGEASVSPEKTSTT | |
| TPR | P12270 | S2155 | GFAEAIHSPQVAGVP | |
| ZNF318 | Q5VUA4 | S2101 | PRSVRIPSPNILKTG | |
| ZNF592 | Q92610 | S322 | TKESSKGSPKMPKSP |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
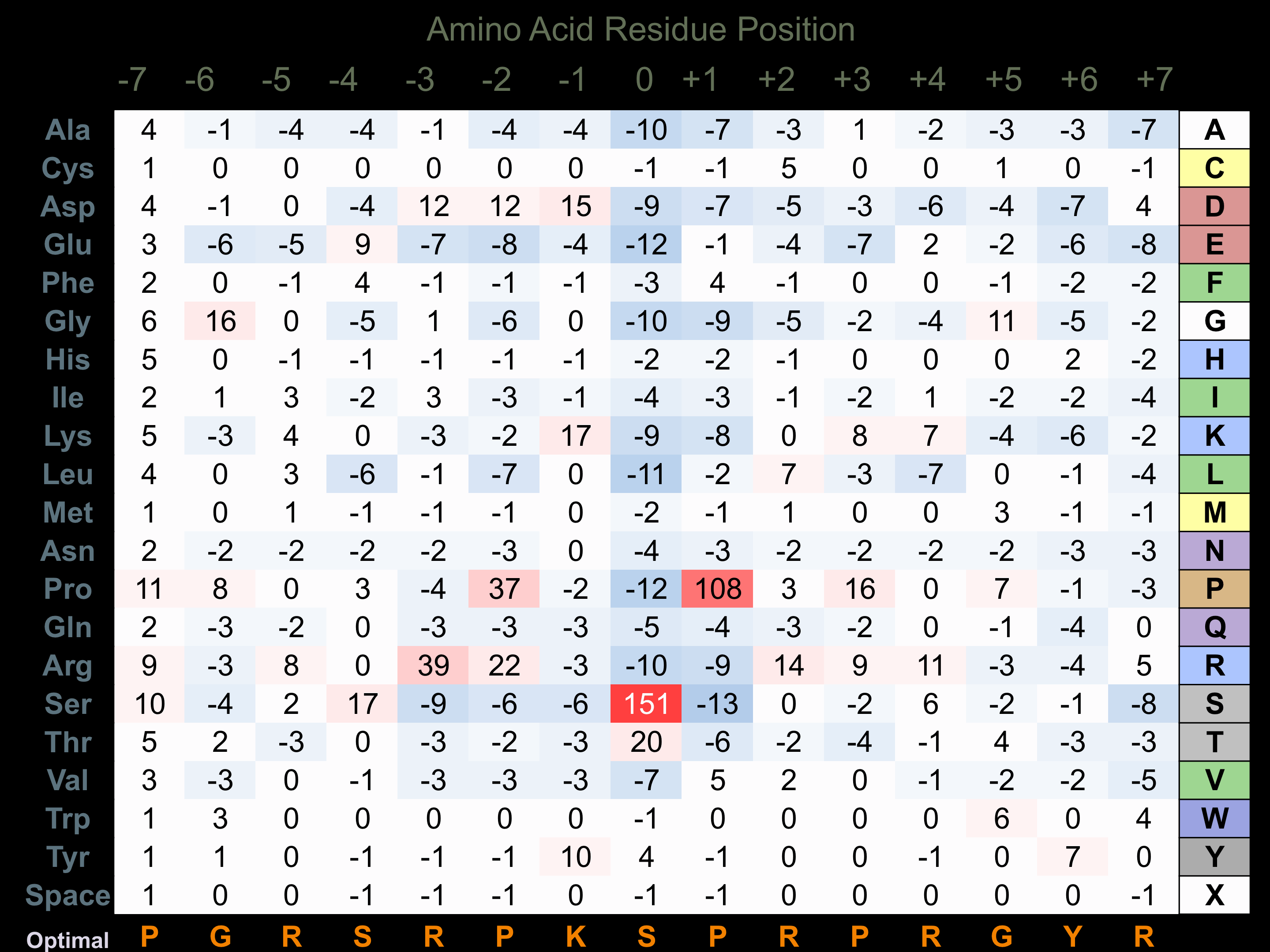
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Nintedanib | Kd = 8.4 nM | 9809715 | 502835 | 22037378 |
| N-Benzoylstaurosporine | Kd = 55 nM | 56603681 | 608533 | 22037378 |
| Staurosporine | Kd = 220 nM | 5279 | 22037378 | |
| KW2449 | Kd = 270 nM | 11427553 | 1908397 | 22037378 |
| Sunitinib | Kd = 390 nM | 5329102 | 535 | 22037378 |
| TG101348 | Kd = 390 nM | 16722836 | 1287853 | 22037378 |
| Dovitinib | Kd = 560 nM | 57336746 | 22037378 | |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| SU14813 | Kd = 1.3 µM | 10138259 | 1721885 | 22037378 |
| Ruboxistaurin | Kd = 1.6 µM | 153999 | 91829 | 22037378 |
| Tozasertib | Kd = 2.5 µM | 5494449 | 572878 | 22037378 |
| PHA-665752 | Kd = 2.6 µM | 10461815 | 450786 | 22037378 |
| A674563 | Kd = 4.6 µM | 11314340 | 379218 | 22037378 |
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Pancreatic cancer; Ovarian cancer; Breast cancer; Colon cancer; Large B-cell lymphoma
Comments:
The enzymatic activity of PRP4 is required for viability of pancreatic, colon and large B-cell lymphoma cancer cell lines (PubMed: 24003220). Inhibition of PRP4 activity by shRNAs was shown to resensitize chemoresistant human ovarian and breast cancer to paclitaxel treatment.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in human Bladder carcinomas (%CFC= +164, p<0.0001); Brain glioblastomas (%CFC= -59, p<0.0001); Brain oligodendrogliomas (%CFC= -83, p<0.0001); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= -95, p<0.0001); Malignant pleural mesotheliomas (MPM) tumours (%CFC= +52, p<0.061); Uterine leiomyomas (%CFC= -74, p<0.021); and Uterine leiomyosarcomas (%CFC= -49, p<0.068).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 24888 diverse cancer specimens. This rate is only -15 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.13 % in 1119 large intestine cancers tested; 0.1 % in 1270 liver cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,561 cancer specimens
Comments:
Only 6 deletions (all at K269fs*13), 1 insertion, and no complex mutations are noted on the COSMIC website.

