Nomenclature
Short Name:
PSKH1
Full Name:
Serine-threonine-protein kinase H1
Alias:
- PSK-H1
- HUMPSKB
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
PSK
SubFamily:
NA
Structure
Mol. Mass (Da):
48035
# Amino Acids:
424
# mRNA Isoforms:
1
mRNA Isoforms:
48,035 Da (424 AA; P11801)
4D Structure:
Homodimer.
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 98 | 355 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Myristoylated:
G2.
Palmitoylated:
C3, C5.
Threonine phosphorylated:
T164, T176.
Tyrosine phosphorylated:
Y35.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 87
87
762
16
996
 9
9
79
11
65
 8
8
74
1
0
 65
65
571
53
1099
 79
79
690
14
668
 43
43
374
45
814
 78
78
683
27
742
 49
49
426
28
570
 37
37
323
10
295
 31
31
273
72
225
 15
15
132
22
116
 53
53
463
130
546
 9
9
83
12
16
 6
6
56
14
44
 20
20
178
20
124
 8
8
69
9
32
 8
8
73
272
53
 8
8
73
12
47
 11
11
98
55
46
 60
60
523
56
545
 15
15
135
19
110
 17
17
145
20
104
 13
13
112
2
13
 11
11
100
12
92
 13
13
113
18
109
 100
100
877
31
1307
 8
8
73
15
45
 7
7
62
10
52
 7
7
58
12
47
 58
58
512
14
77
 82
82
720
18
489
 53
53
467
27
659
 42
42
364
46
607
 97
97
851
26
717
 13
13
116
22
104
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 58.2
58.2
72.6
100 99.7
99.7
99.7
100 -
-
-
98 -
-
-
96 55.6
55.6
71.9
97 -
-
-
- 97.6
97.6
98.3
98 37
37
54.2
97 -
-
-
- 56.3
56.3
70
- -
-
-
89 32.5
32.5
54.2
84 80.1
80.1
87.2
82 -
-
-
- -
-
-
- 37.7
37.7
56.1
- 39.3
39.3
59.4
47 45
45
57.7
- -
-
-
- -
-
-
- -
-
-
38 -
-
-
39 -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CDC6 - Q99741 |
Regulation
Activation:
Activated by Ca2+.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
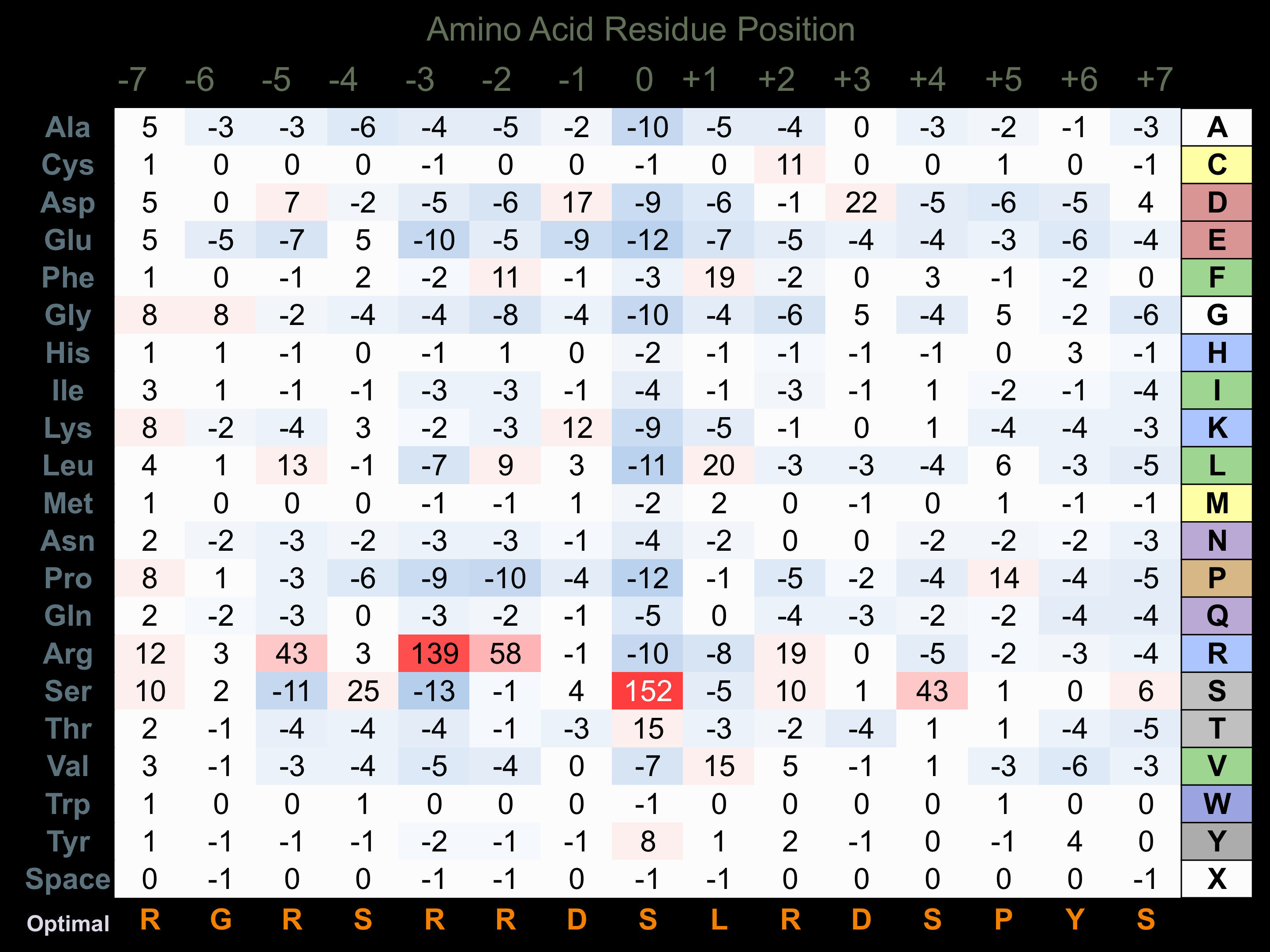
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Inflammatory disorders
Specific Diseases (Non-cancerous):
Crohn's disease
Comments:
PSKH1 is reported to be upregulated in Crohn's disease.
Comments:
PSKH1 has been identified as a regulator of prostate cancer cell growth.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for PSKH1 in diverse human cancers of 396, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 166 for this protein kinase in human cancers was 2.8-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. A D218A substitution in PSKH1 is associated with loss of its autophosphorylation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 24726 diverse cancer specimens. This rate is only -24 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.4 % in 589 stomach cancers tested; 0.23 % in 603 endometrium cancers tested; 0.19 % in 1270 large intestine cancers tested; 0.11 % in 864 skin cancers tested; 0.1 % in 1634 lung cancers tested; 0.09 % in 1512 liver cancers tested; 0.08 % in 942 upper aerodigestive tract cancers tested; 0.07 % in 710 oesophagus cancers tested; 0.07 % in 1316 breast cancers tested; 0.06 % in 1276 kidney cancers tested; 0.04 % in 548 urinary tract cancers tested; 0.03 % in 1459 pancreas cancers tested; 0.01 % in 2082 central nervous system cancers tested; 0.01 % in 2009 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R369H (3).
Comments:
Only 3 deletions, and no insertions or complex mutations are noted on the COSMIC website.

