Nomenclature
Short Name:
CYGD
Full Name:
Retinal guanylyl cyclase 1
Alias:
- Guanylate cyclase 2D, retinal
- RETGC-1
- Rod outer segment membrane guanylate cyclase
- ROS-GC
- GUCY2D
Classification
Type:
Protein-serine/threonine kinase
Group:
RGC
Family:
RGC
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
120,059
# Amino Acids:
1103
# mRNA Isoforms:
1
mRNA Isoforms:
120,059 Da (1103 AA; Q02846)
4D Structure:
NA
1D Structure:
Subfamily Alignment
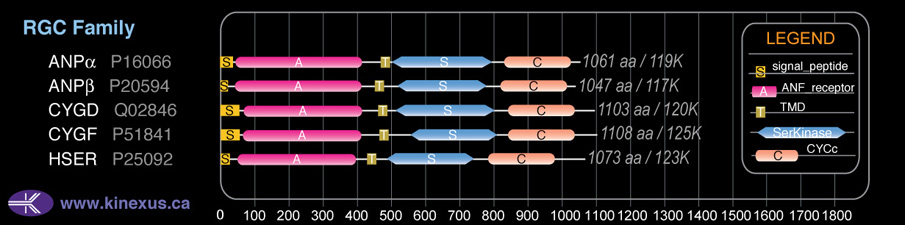
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 1 | 51 | signal_peptide |
| 72 | 409 | ANF_receptor |
| 465 | 487 | TMD |
| 534 | 817 | Pkinase |
| 844 | 1039 | CYCc |
| 880 | 1010 | Guanylate cyclase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
N-GlcNAcylated:
N297.
Serine phosphorylated:
S200, S214, S298, S527, S529, S535, S645.
Tyrosine phosphorylated:
Y283.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 38
38
822
13
1360
 0.7
0.7
16
9
18
 7
7
152
20
371
 8
8
165
59
315
 11
11
239
14
233
 0.6
0.6
13
37
7
 15
15
332
19
642
 14
14
301
34
483
 8
8
166
10
158
 4
4
85
60
108
 2
2
54
33
69
 20
20
436
111
543
 3
3
61
29
59
 0.9
0.9
20
6
22
 6
6
121
30
212
 0.7
0.7
16
8
21
 3
3
66
115
68
 4
4
82
26
99
 2
2
51
62
34
 11
11
231
56
285
 4
4
97
25
218
 2
2
50
27
77
 3
3
62
28
79
 3
3
55
25
69
 3
3
64
26
71
 14
14
296
41
425
 2
2
47
34
55
 3
3
65
23
75
 6
6
121
25
353
 6
6
138
14
64
 18
18
396
18
285
 100
100
2171
21
4461
 7
7
154
48
474
 24
24
521
31
537
 8
8
174
22
133
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
99.5 97.5
97.5
98.3
98 -
-
-
87 -
-
-
- 81.8
81.8
86.9
87 -
-
-
- 85.1
85.1
90.1
85 84.9
84.9
89.7
85 -
-
-
- 50.9
50.9
66.1
- 53.1
53.1
69.6
60 -
-
-
- 50.6
50.6
68
55 -
-
-
- 28.3
28.3
47.1
- -
-
-
- 26.9
26.9
44.4
- 31.3
31.3
47.2
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Activated by GCAP-1
Inhibition:
Inhibited by calcium.
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
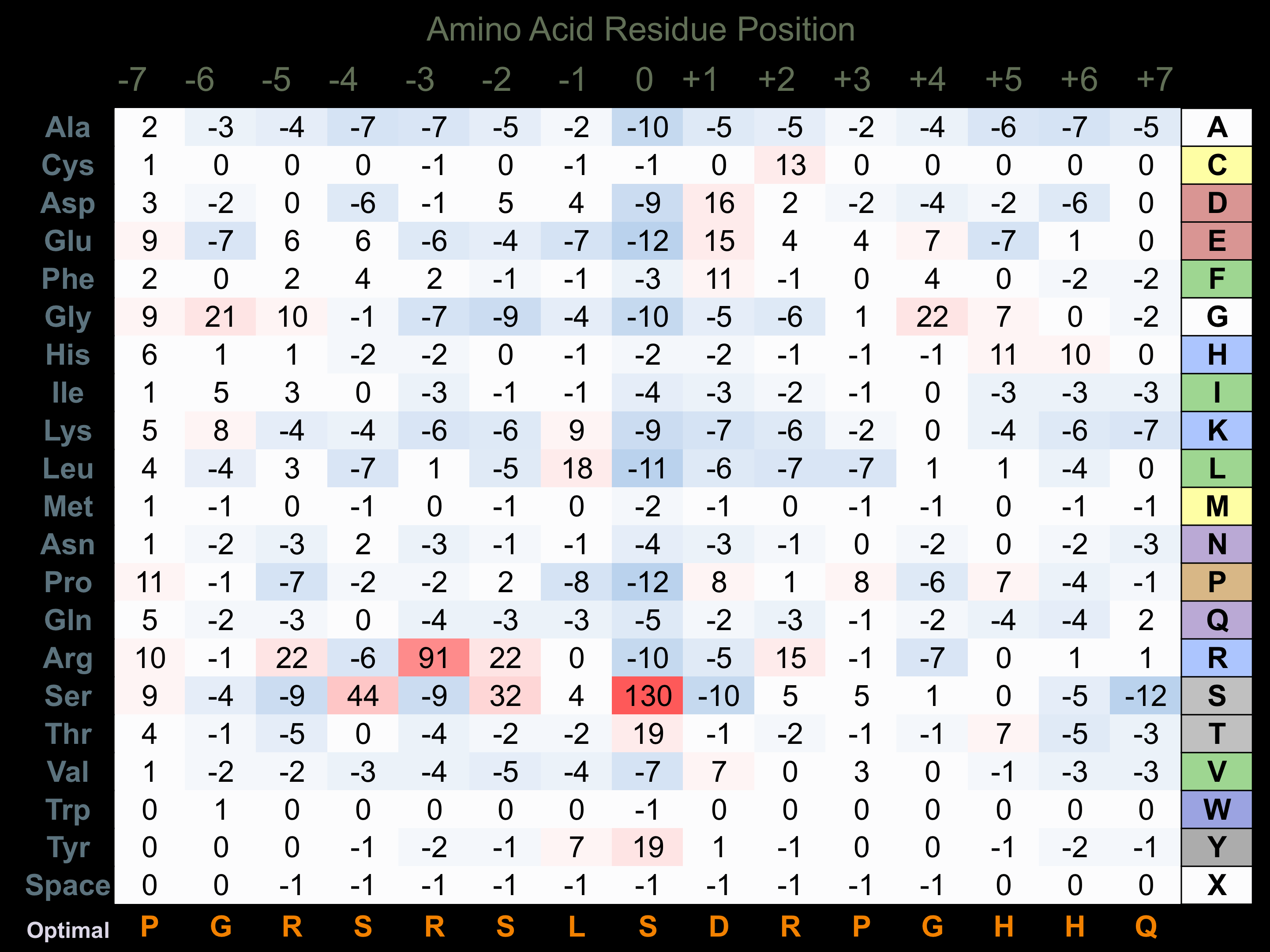
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Eye disorders
Specific Diseases (Non-cancerous):
Cone-rod dystrophy; Leber congenital amaurosis; Cone-rod dystrophy 6; Keratoconus; Leber congenital amaurosis 1; Rpe65-related Leber congenital amaurosis; Aipl1-related Leber congenital amaurosis; Partial central choroid dystrophy; Hyperopia; Cone-rod dystrophy 2; Leber congenital amaurosis 17; Choriodal dystrophy, Central areolar 2; Gucy2d-related Leber congenital amaurosis
Comments:
Cone-rod dystrophies are inherited eye diseases that affect the rod and cone cells (photoreceptor cells) of the retina. In this disease, deterioration of the cones is more pronounced than the rods, resulting in more advanced central vision loss, loss of normal colour perception, and abnormal sensitivity to light (photophobia), followed by progressive peripheral vision loss and night blindness. This disease can be inherited in an autosomal recessive, autosomal dominant, X-linked, or maternally inherited manner and has been linked to mutations in several genes. For example, several loss-of-function mutations in the CYGD gene have been observed in patients with cone-rod dystrophies, including the R838C, E837D, T839M, and I949T substitution mutations. These mutations are predicted to decrease the kinase catalytic activity of the protein, however they may not completely abolish it. Leber congential amaurosis is a congenital eye disease that affects the retina, causing severe visual defects, combined with photophobia, involuntary eye movement (nystagmus), and farsightedness. Several loss-of-function mutations in the CYGD gene have been identified in patients with Leber congenital amaurosis including a F565S substitution mutation that significantly reduces the kinase catalytic activity of the protein, a deletion mutation that removes either 460C or 693C causing a frameshift mutation and premature stop codon, and a A52S substitution mutation. The effect of the A52S mutation is unclear as it was difficult to discern the effect on the kinase catalytic activity.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 24447 diverse cancer specimens. This rate is only -20 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.4 % in 864 skin cancers tested; 0.27 % in 1229 large intestine cancers tested; 0.24 % in 569 stomach cancers tested; 0.11 % in 603 endometrium cancers tested; 0.1 % in 548 urinary tract cancers tested; 0.1 % in 273 cervix cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: Q694H (4).
Comments:
Only 1 complex mutation and no deletions or insertions noted on the COSMIC website.

