Nomenclature
Short Name:
RNASEL
Full Name:
2-5A-dependent ribonuclease
Alias:
- 2-5A-dependent ribonuclease
- 2-5A-dependent RNase
- RN5A
- RNase L
- RNS4
- EC 3.1.26.-
- PRCA1
- Ribonuclease 4
- Ribonuclease L
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
Other-Unique
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
83,533
# Amino Acids:
741
# mRNA Isoforms:
2
mRNA Isoforms:
83,533 Da (741 AA; Q05823); 73,416 Da (652 AA; Q05823-2)
4D Structure:
Monomer (inactive form) or homodimer. Interacts with ABCE1; this interaction inhibits the RNASEL
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
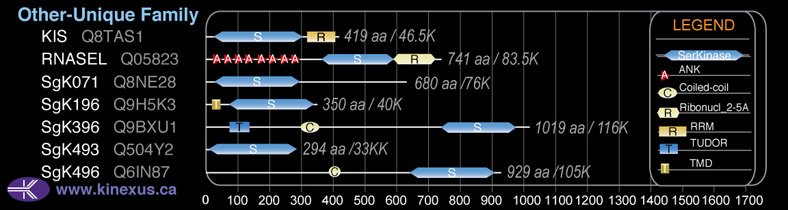
Domain Distribution:
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K684 (N6).
Serine phosphorylated:
S15, S16, S17.
Ubiquitinated:
K287.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 96
96
1063
25
1199
 4
4
49
9
34
 4
4
47
6
37
 43
43
471
72
1470
 52
52
575
24
495
 2
2
25
46
18
 8
8
86
25
73
 38
38
417
21
602
 20
20
220
10
196
 5
5
58
62
41
 4
4
48
17
45
 72
72
797
74
615
 6
6
68
17
43
 3
3
38
6
28
 4
4
44
9
46
 4
4
40
12
22
 9
9
96
83
79
 4
4
49
11
33
 2
2
23
60
20
 50
50
547
84
545
 5
5
60
13
42
 10
10
115
15
135
 5
5
58
15
58
 2
2
20
11
20
 14
14
151
13
214
 60
60
668
47
648
 5
5
54
20
29
 5
5
53
11
44
 6
6
61
11
42
 6
6
61
28
59
 100
100
1105
18
749
 28
28
310
32
457
 11
11
125
29
82
 88
88
973
57
831
 7
7
72
35
74
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 97.3
97.3
98.8
97 91
91
94.5
92.5 -
-
-
72 -
-
-
- -
-
-
71.5 -
-
-
- 64.1
64.1
77.2
65 62.8
62.8
79.1
64 -
-
-
- -
-
-
- 22.1
22.1
36.6
44 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | ABCE1 - P61221 |
Regulation
Activation:
After binding to 2-5A (5'-phosphorylated 2',5'-linked oligoadenylates) the homodimerization and subsequent activation occurs.
Inhibition:
Inhibited by RNASEL inhibitor ABCE1/RLI, a cytoplasmic member of the ATP-binding cassette (ABC) transporter family.
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
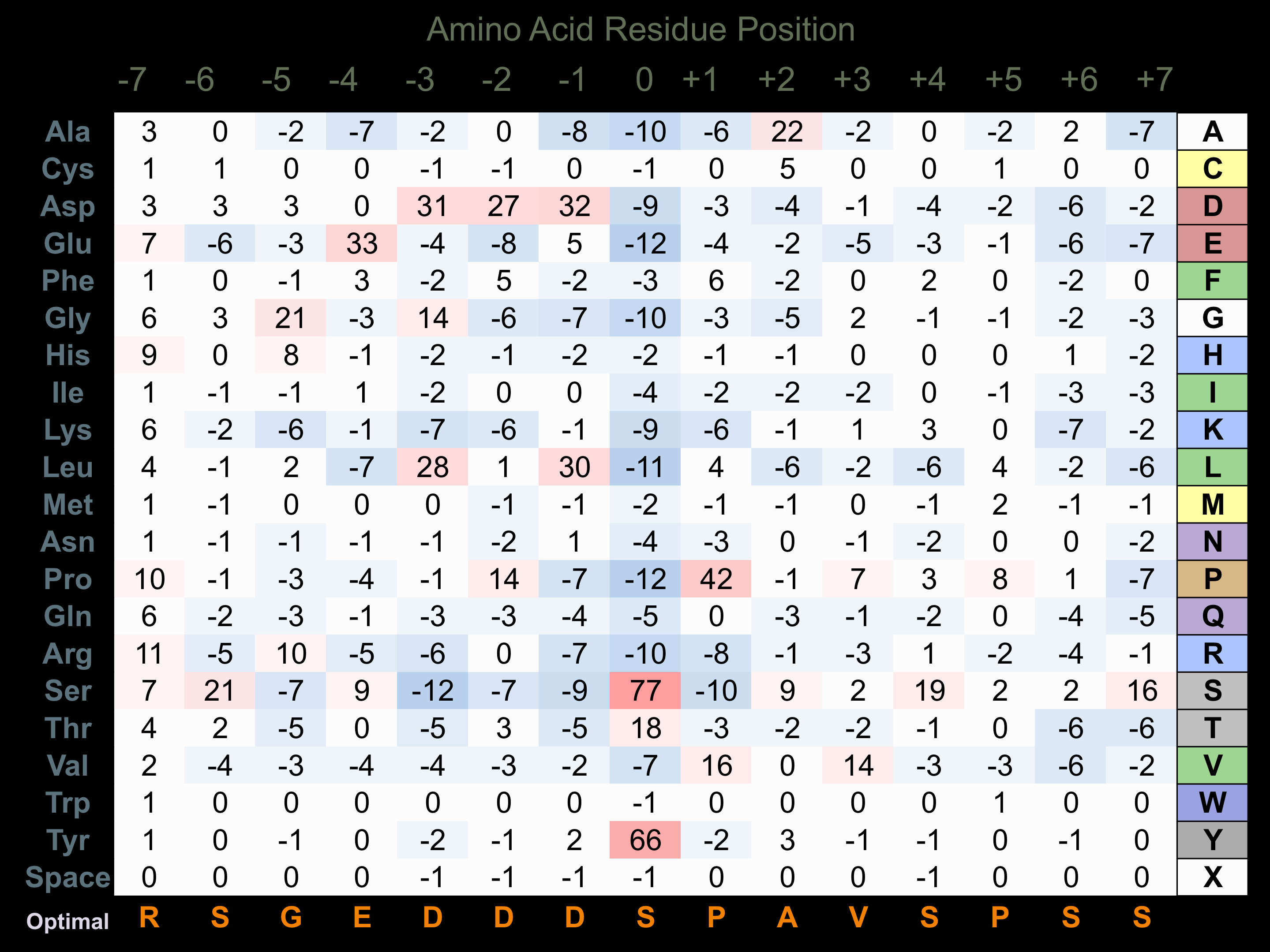
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Cancer, neurological, and endocrine disorders
Specific Diseases (Non-cancerous):
Chronic fatigue syndrome (CFS); Parainfluenza virus Type 3
Comments:
Chronic Fatigue Syndrome (CFS) is a rare neuronal disorder characterized by debilitating, long-lasting fatigue which prevents normal day-to-day functioning. CFS can also be characterized by muscle pain after exercise, memory impairment, headaches, joint pains, sleep issues, sore throat, and sore lymph nodes. Parainfluenza Virus Type 3 is a rare condition with a relation to cytokine, inflammatory, immune, and MIF-mediated glucocorticoid regulation pathways. The K240N and K274N mutations in conjunction lead to almost full inhibition of 2-5A binding. RNASEL enzyme activity and dimerization is inhibited with a K392R mutation, whereas just enzyme activity is impaired with a D661A, or R667A, or H672A mutation.
Specific Cancer Types:
Prostate cancer 1; Prostate cancer; Familial prostate cancer; Prostatitis
Comments:
RNASEL is linked to Prostate cancer 1, which is a rare condition that is related to the interferon signalling pathway and is related to the prostatitis and Prostate Cancer conditions. Familial Prostate Cancer is a rare cancer related to prostate cancer and to breast cancer. Prostate Cancer, Progression and Metastasis is a rare condition linked to pathways that include the response to DNA damage and the ATM pathway. The K240N and K274N mutations in conjunction lead to almost full inhibition of 2-5A binding. RNASEL enzyme activity and dimerization is inhibited with a K392R mutation, whereas just enzyme activity is impaired with a D661A, or R667A, or H672A mutation.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for RNASEL in diverse human cancers of 598, which is 1.3-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 112 for this protein kinase in human cancers was 1.9-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 24726 diverse cancer specimens. This rate is very similar (+ 8% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.9 % in 15 pituitary cancers tested; 0.41 % in 864 skin cancers tested; 0.36 % in 603 endometrium cancers tested; 0.31 % in 1270 large intestine cancers tested; 0.25 % in 589 stomach cancers tested; 0.15 % in 1634 lung cancers tested; 0.11 % in 127 biliary tract cancers tested; 0.1 % in 548 urinary tract cancers tested; 0.1 % in 273 cervix cancers tested; 0.08 % in 833 ovary cancers tested; 0.08 % in 710 oesophagus cancers tested; 0.08 % in 1512 liver cancers tested; 0.06 % in 1276 kidney cancers tested; 0.05 % in 558 thyroid cancers tested; 0.04 % in 1316 breast cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: D687G (3).
Comments:
Only 3 deletions, 3 insertions, and no complex mutations are noted on the COSMIC website.

