Nomenclature
Short Name:
DLK
Full Name:
Mitogen-activated protein kinase kinase kinase 12
Alias:
- Dual leucine zipper bearing kinase
- Dual leucine zipper kinase DLK
- MAP3K12
- MAPK-upstream kinase
- MEKK12
- ZPKP1
- EC 2.7.11.25
- Kinase DLK
- Leucine-zipper protein kinase
- M3K12
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
Family:
MLK
SubFamily:
LZK
Specific Links
Structure
Mol. Mass (Da):
93219
# Amino Acids:
859
# mRNA Isoforms:
2
mRNA Isoforms:
96,322 Da (892 AA; Q12852-2); 93,219 Da (859 AA; Q12852)
4D Structure:
Interacts with MBIP
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 125 | 366 | Pkinase |
| 419 | 467 | Coiled-coil |
| 390 | 411 | Leucine zipper 1 |
| 443 | 464 | Leucine zipper 2 |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S615, S634, S645, S695, S757.
Threonine phosphorylated:
T642, T699.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 35
35
962
32
1180
 2
2
57
15
46
 8
8
233
16
564
 15
15
425
95
581
 22
22
600
25
500
 2
2
62
80
153
 8
8
227
31
535
 17
17
461
43
625
 10
10
269
17
195
 6
6
152
87
629
 5
5
125
35
391
 26
26
708
166
710
 4
4
98
38
291
 3
3
76
12
34
 7
7
182
29
625
 2
2
66
15
37
 3
3
72
126
133
 6
6
172
25
566
 3
3
85
91
366
 15
15
400
109
446
 8
8
212
27
507
 5
5
129
31
411
 4
4
112
26
244
 7
7
198
25
721
 9
9
244
27
794
 19
19
511
61
654
 4
4
99
41
374
 6
6
153
25
429
 6
6
176
25
326
 2
2
63
28
35
 28
28
765
24
710
 100
100
2744
36
6410
 0.8
0.8
23
65
96
 27
27
731
57
667
 3
3
90
35
98
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 95.8
95.8
95.9
100 95.3
95.3
95.6
99 -
-
-
97 -
-
-
92 97.4
97.4
98.6
97.5 -
-
-
- 92.6
92.6
93.8
96 91.8
91.8
93.1
96 -
-
-
- -
-
-
- 52.4
52.4
62.5
- 51.8
51.8
63.2
74 65.4
65.4
74
78 -
-
-
- 24
24
38.4
- 38.4
38.4
52.9
- 29.1
29.1
44.9
- 38.6
38.6
52.8
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | MAPK8IP1 - Q9UQF2 |
| 2 | MBIP - Q9NS73 |
| 3 | TGM2 - P21980 |
| 4 | MAP3K11 - Q16584 |
| 5 | MAP2K4 - P45985 |
| 6 | SH3RF1 - Q7Z6J0 |
| 7 | MAPK8IP2 - Q13387 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
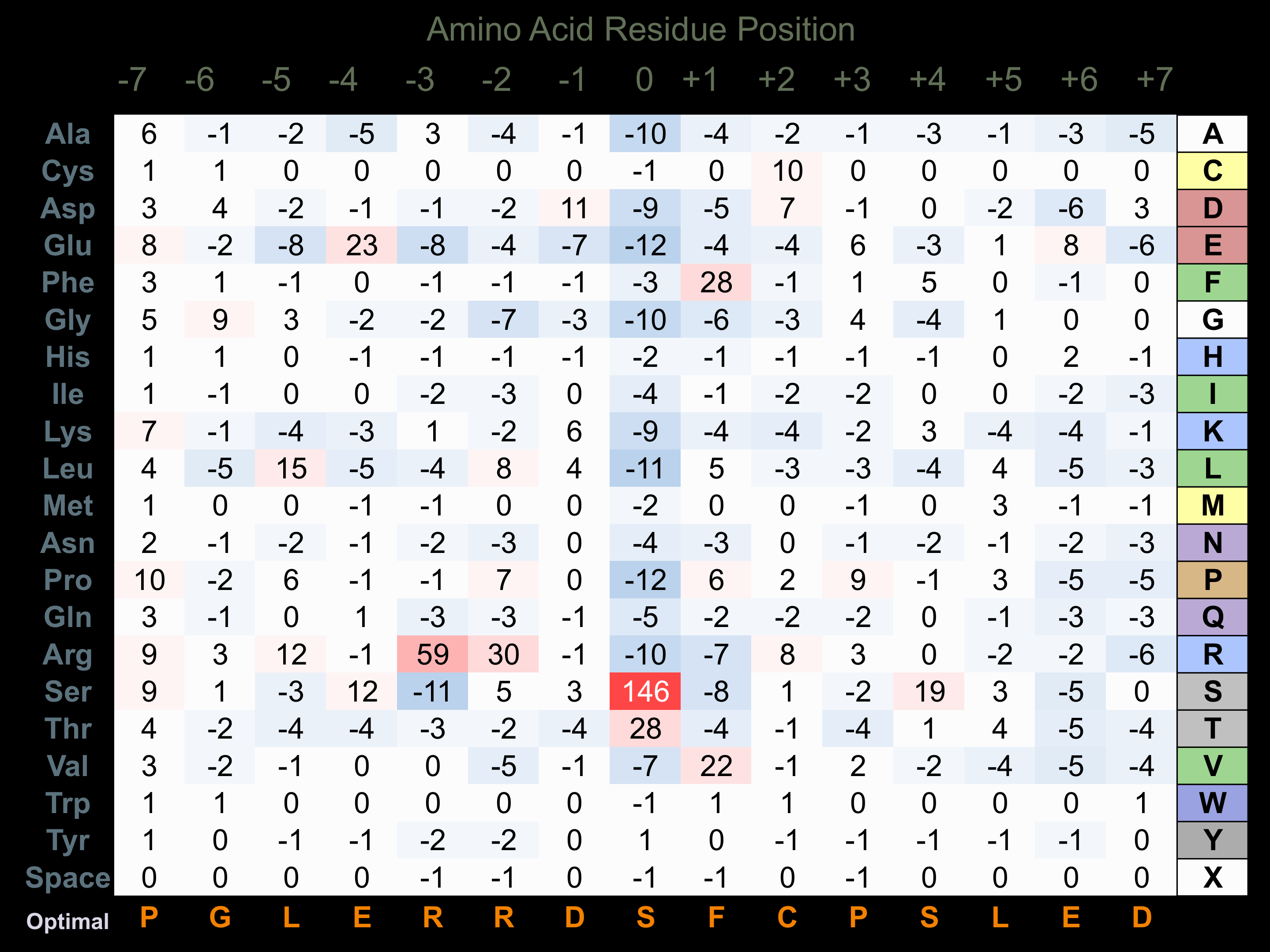
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Malignant pleural mesotheliomas (MPM) tumours (%CFC= +112, p<0.001); Ovary adenocarcinomas (%CFC= +52, p<0.056); Skin melanomas - malignant (%CFC= +210, p<0.0001); and Uterine leiomyomas (%CFC= -45, p<0.013). The COSMIC website notes an up-regulated expression score for DLK in diverse human cancers of 394, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 7 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24995 diverse cancer specimens. This rate is only -1 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.39 % in 1093 large intestine cancers tested; 0.28 % in 589 stomach cancers tested; 0.27 % in 805 skin cancers tested; 0.12 % in 1270 liver cancers tested; 0.09 % in 1807 lung cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,279 cancer specimens
Comments:
Twelve deletions, 1 insertion and no complex mutations are noted on the COSMIC website.

