Nomenclature
Short Name:
PDHK3
Full Name:
[Pyruvate dehydrogenase [lipoamide]] kinase isozyme 3, mitochondrial
Alias:
- EC 2.7.11.2
- Kinase Pyruvate dehydrogenase kinase 3
- PDK3
Classification
Type:
Protein-serine/threonine kinase
Group:
Atypical
Family:
PDHK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
46,939
# Amino Acids:
406
# mRNA Isoforms:
2
mRNA Isoforms:
48,043 Da (415 AA; Q15120-2); 46,939 Da (406 AA; Q15120)
4D Structure:
Interacts with the pyruvate dehydrogenase complex E2 subunit (DLAT).
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
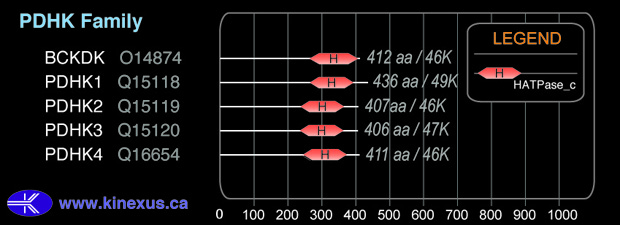
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 236 | 362 | HATPase_c |
| 131 | 362 | Histidine kinase |
| 25 | 189 | BCDHK |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K30, K296.
Serine phosphorylated:
S239.
Threonine phosphorylated:
T169.
Tyrosine phosphorylated:
Y19.
Ubiquitinated:
K30.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 59
59
1225
54
1239
 2
2
44
25
59
 6
6
119
13
150
 16
16
331
188
455
 44
44
918
56
737
 3
3
59
134
98
 13
13
265
71
430
 33
33
684
65
1221
 26
26
530
24
402
 8
8
174
191
219
 5
5
96
49
175
 27
27
552
293
594
 3
3
66
46
81
 2
2
44
21
46
 3
3
68
28
110
 2
2
36
34
38
 2
2
44
315
91
 8
8
174
28
261
 3
3
57
169
58
 33
33
690
218
677
 4
4
90
37
122
 5
5
103
44
124
 4
4
79
23
115
 8
8
158
28
164
 6
6
117
38
133
 61
61
1264
114
2865
 3
3
63
49
70
 6
6
132
28
221
 3
3
69
26
87
 6
6
125
70
109
 26
26
531
30
613
 100
100
2068
61
5127
 7
7
137
114
255
 47
47
980
135
862
 18
18
368
74
352
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 63.5
63.5
77.8
100 -
-
-
100 -
-
-
98 -
-
-
- 97.1
97.1
97.3
98.5 -
-
-
- 95.2
95.2
96.6
97 64.1
64.1
78.3
97 -
-
-
- 63.8
63.8
78.6
- 92.9
92.9
96.5
93 69.2
69.2
81.8
85 82
82
89.7
82 -
-
-
- 55.9
55.9
70.2
60 -
-
-
- 51.5
51.5
66.3
54 -
-
-
- -
-
-
- -
-
-
- -
-
-
43 -
-
-
42 -
-
-
- -
-
-
47
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | DLAT - P10515 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Disease Linkage
General Disease Association:
Cancer, musculoskeletal, and neuronal disorders
Specific Diseases (Non-cancerous):
Charcot-Marie-Tooth disease, X-linked dominant, 6
Comments:
An R158H substitution in PDHK2 is associated with charcot-Marie-Tooth disease, X-linked dominant, 6, which is a disorder of the peripheral nervous system, featured by weakness and atrophy, initially of the peroneal muscles and later of the distal muscles of the arms.
Specific Cancer Types:
Colon cancer
Comments:
Inactivation of PDHK3 signalling drives melanomas towards oxidative metabolism and allows for the possibility of using pro-oxidants as therapies. PDHK3 has been shown to be overexpressed in colon cancer cell lines.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= +354, p<0.0007); Brain oligodendrogliomas (%CFC= +502, p<(0.0003); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +981, p<0.0001); Ovary adenocarcinomas (%CFC= -49, p<0.071); and Pituitary adenomas (aldosterone-secreting) (%CFC= +95, p<0.003). The COSMIC website notes an up-regulated expression score for PDHK3 in diverse human cancers of 671, which is 1.5-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 257 for this protein kinase in human cancers was 4.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 24752 diverse cancer specimens. This rate is very similar (+ 6% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 1.64 % in 15 pituitary cancers tested; 0.53 % in 603 endometrium cancers tested; 0.39 % in 1270 large intestine cancers tested; 0.21 % in 589 stomach cancers tested; 0.18 % in 273 cervix cancers tested; 0.18 % in 1634 lung cancers tested; 0.14 % in 864 skin cancers tested; 0.1 % in 958 upper aerodigestive tract cancers tested; 0.1 % in 1512 liver cancers tested; 0.06 % in 881 prostate cancers tested; 0.06 % in 833 ovary cancers tested; 0.05 % in 1459 pancreas cancers tested.
Frequency of Mutated Sites:
None > 6 in 20,035 cancer specimens
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.

