Nomenclature
Short Name:
TRKC
Full Name:
NT-3 growth factor receptor
Alias:
- EC 2.7.10.1
- GP145-TrkC
- NT-3 growth factor receptor
- NTRK3
- TRKC
- TrkC tyrosine kinase
- Kinase TrkC
- Neurotrophic tyrosine kinase receptor 3
- Neurotrophic tyrosine kinase, receptor, type 3
- Neurotrophin-3 receptor non-catalytic isoform 1
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Trk
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
94428
# Amino Acids:
839
# mRNA Isoforms:
5
mRNA Isoforms:
94,428 Da (839 AA; Q16288); 93,460 Da (831 AA; Q16288-4); 92,801 Da (825 AA; Q16288-3); 91,833 Da (817 AA; Q16288-5); 68,452 Da (612 AA; Q16288-2)
4D Structure:
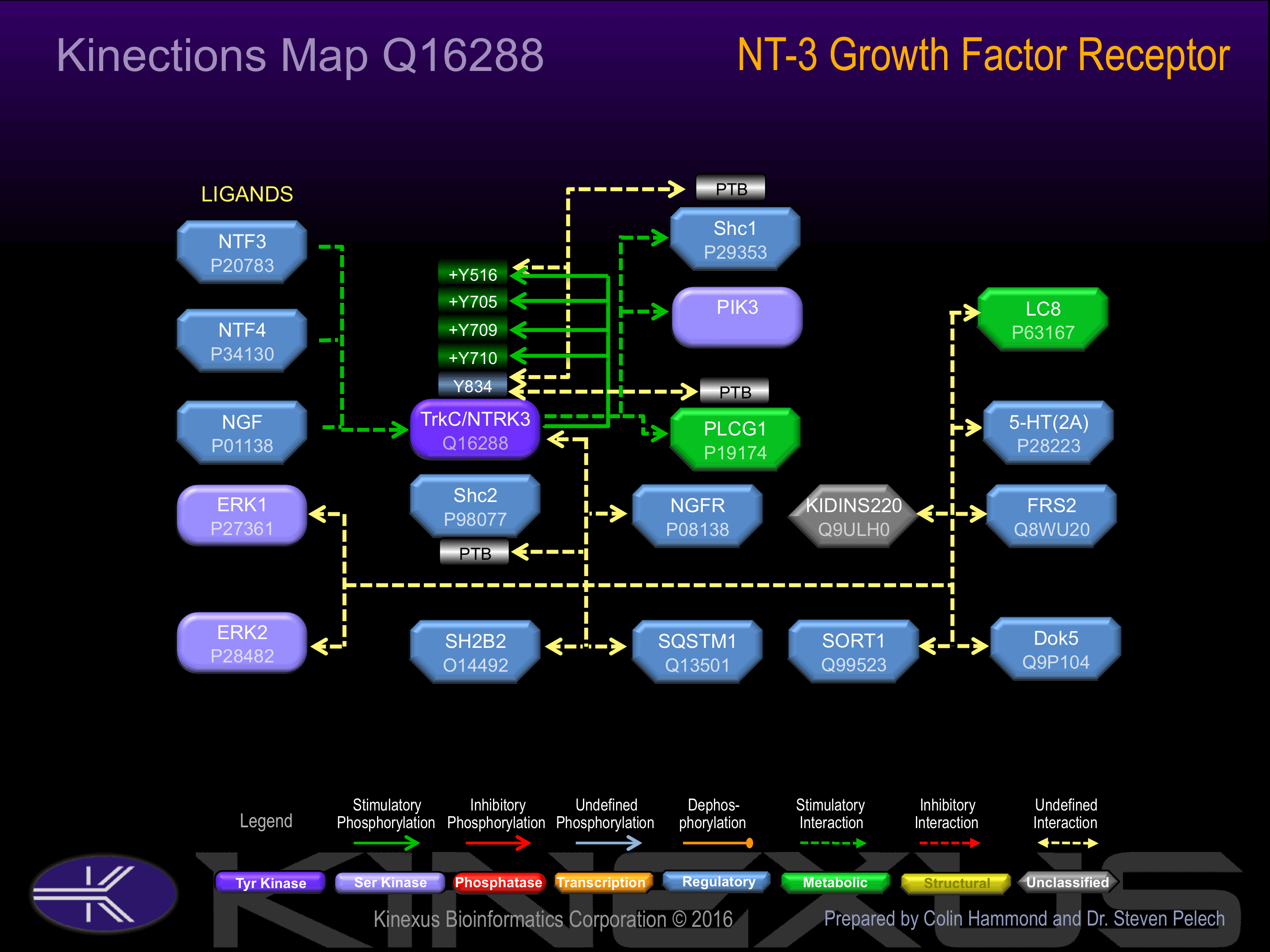
Exists in a dynamic equilibrium between monomeric (low affinity) and dimeric (high affinity) structures. Binds SH2B2. Interacts with SQSTM1 and KIDINS220
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
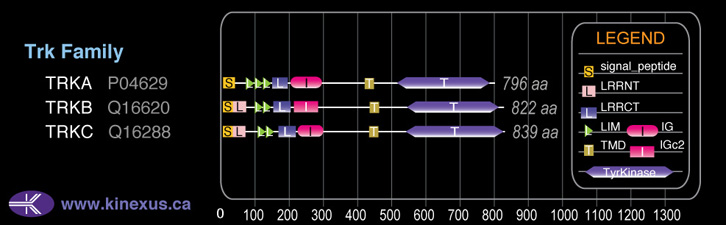
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
N-GlcNAcylated:
N72, N79, N133, N163, N203, N218, N232, N259, N267, N272, N294, N375, N388.
Serine phosphorylated:
S701, S706.
Threonine phosphorylated:
T220, T386.
Tyrosine phosphorylated:
Y131, Y516+, Y558, Y705+, Y709+, Y710+, Y800, Y834.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1060
106
1223
 7
7
76
52
88
 7
7
72
23
72
 45
45
479
335
559
 43
43
461
99
350
 2
2
21
277
19
 18
18
196
130
441
 52
52
546
112
633
 26
26
280
55
178
 9
9
96
311
119
 7
7
76
90
94
 60
60
641
586
614
 5
5
50
101
70
 8
8
81
39
73
 9
9
99
54
168
 3
3
32
63
29
 4
4
40
529
61
 8
8
85
50
94
 7
7
78
309
65
 29
29
306
428
304
 18
18
186
66
294
 6
6
67
76
115
 4
4
45
47
58
 9
9
95
54
109
 7
7
69
63
108
 50
50
532
195
582
 5
5
50
103
51
 9
9
93
51
112
 8
8
84
52
80
 40
40
422
112
412
 22
22
228
66
290
 69
69
736
121
1272
 6
6
61
170
273
 73
73
777
244
705
 25
25
264
126
204
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 98.1
98.1
98.2
100 99.4
99.4
99.9
99 -
-
-
99 -
-
-
96 88
88
89.5
98 -
-
-
- 95.7
95.7
97.4
97 94.6
94.6
96.2
97 -
-
-
- 65.2
65.2
74.1
- 88.7
88.7
93.8
89 56.5
56.5
70.1
85 64.5
64.5
71.4
76 -
-
-
- 26.2
26.2
43.6
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | NGFR - P08138 |
| 2 | DOK5 - Q9P104 |
| 3 | MAPK1 - P28482 |
| 4 | SHC2 - P98077 |
| 5 | MAPK3 - P27361 |
| 6 | DYNLL1 - P63167 |
| 7 | KIDINS220 - Q9ULH0 |
| 8 | FRS2 - Q8WU20 |
| 9 | BDNF - P23560 |
Regulation
Activation:
Phosphorylation of Tyr-516 increases phosphotransferase activity. Phosphorylation of Tyr-834 induces interaction with PLCg1 and Shc1.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
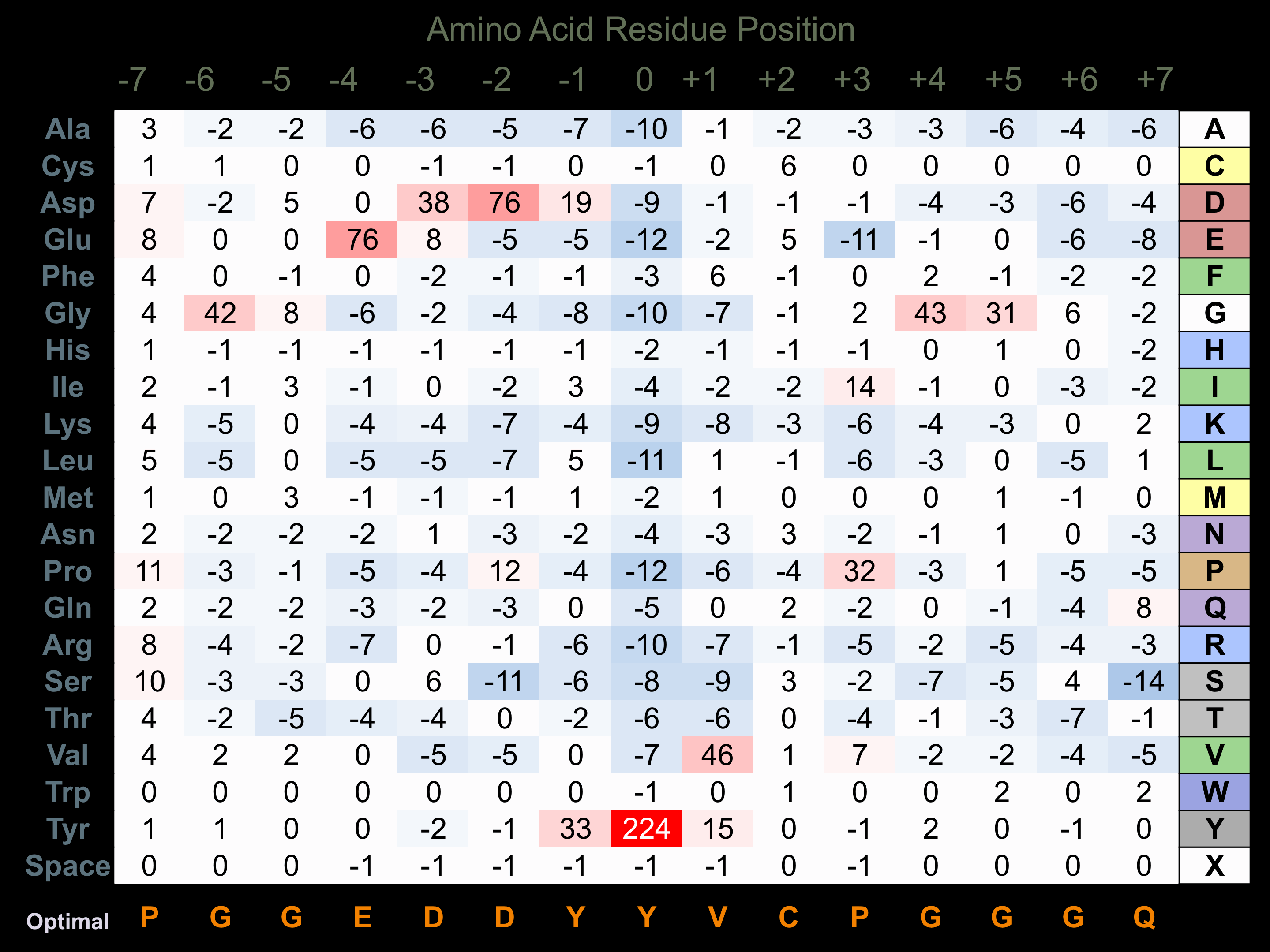
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Fibrosarcomas; medulloblastoma; Congenital mesoblastic nephromas (CMN); Mesoblastic nephromas; Congenital fibrosarcomas (CFS); Gastrointestinal stromal tumours (GIST); Polymorphous low-grade adenocarcinomas (PLGA); Medulloblastomas, desmoplastic
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= -62, p<0.0001); Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= -46, p<0.076); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +253, p<0.0002); Malignant pleural mesotheliomas (MPM) tumours (%CFC= -47, p<0.0005); Pituitary adenomas (ACTH-secreting) (%CFC= -65, p<0.018); Skin fibrosarcomas (%CFC= +270, p<0.0002); and Skin melanomas - malignant (%CFC= +1652, p<0.0001). The COSMIC website notes an up-regulated expression score for TRKC in diverse human cancers of 302, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 14 for this protein kinase in human cancers was 0.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.17 % in 27504 diverse cancer specimens. This rate is 2.3-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.97 % in 809 skin cancers tested; 0.63 % in 1374 large intestine cancers tested; 0.41 % in 2581 lung cancers tested; 0.3 % in 602 endometrium cancers tested; 0.29 % in 816 stomach cancers tested; 0.24 % in 605 oesophagus cancers tested; 0.11 % in 1278 liver cancers tested; 0.09 % in 1396 kidney cancers tested.
Frequency of Mutated Sites:
None > 8 in 22,538 cancer specimens
Comments:
Only 8 deletions, no insertions and no complex mutations are noted on the COSMIC website.