Nomenclature
Short Name:
PDHK4
Full Name:
Pyruvate dehydrogenase [lipoamide]] kinase isozyme 4, mitochondrial
Alias:
- Pyruvate dehydrogenase kinase isoform 4
- PDK4
Classification
Type:
Protein-serine/threonine kinase
Group:
Atypical
Family:
PDHK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
46,469
# Amino Acids:
411
# mRNA Isoforms:
1
mRNA Isoforms:
46,469 Da (411 AA; Q16654)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
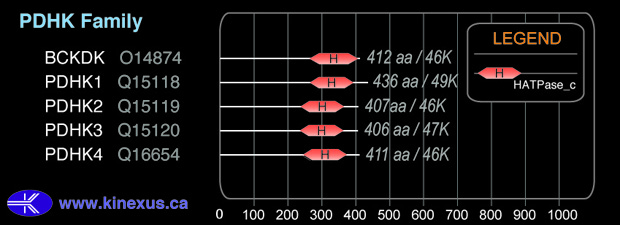
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 243 | 368 | HATPase_c |
| 138 | 368 | Histidine kinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S10, S13, S368, S369, S381.
Ubiquitinated:
K111, K122.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 50
50
1031
30
904
 9
9
192
12
243
 87
87
1771
11
996
 14
14
278
112
644
 57
57
1161
30
775
 2
2
45
59
96
 9
9
184
39
295
 29
29
583
41
672
 36
36
739
10
646
 36
36
725
85
969
 14
14
294
30
551
 37
37
754
133
820
 12
12
244
22
279
 7
7
137
9
88
 10
10
201
17
235
 31
31
632
18
805
 6
6
121
202
223
 11
11
230
18
236
 100
100
2042
78
3301
 37
37
761
107
713
 23
23
461
26
637
 16
16
328
28
424
 29
29
590
20
685
 8
8
154
18
137
 4
4
91
26
95
 47
47
950
74
924
 17
17
337
25
471
 26
26
538
18
526
 19
19
395
18
440
 4
4
81
28
77
 21
21
439
18
338
 12
12
254
31
344
 25
25
515
76
869
 43
43
877
83
818
 83
83
1704
48
4042
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.7
99.7
100
100 98
98
99.5
98 -
-
-
93 -
-
-
97 58.2
58.2
74
94 -
-
-
- 92.7
92.7
96.8
93 92.2
92.2
96.3
93 -
-
-
- 70.3
70.3
74.4
- 77.7
77.7
88.1
80 72
72
85.4
- 64.7
64.7
82.7
73 -
-
-
- 50.6
50.6
68.5
- -
-
-
- 47.6
47.6
65.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | DLAT - P10515 |
| 2 | PDHA1 - P08559 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Disease Linkage
Comments:
Studies in transgenic PDHK4 knockout mice subjected to a high-saturated fat diet that induces hyperglycemia, hyperinsulinaemia, glucose intolerance, hepatic steatosis and obesity, showed that PDHK4 is an attractive candidate for drug discovery for the treatment of type 2 diabetes. Cells that lack insulin (or are insensitive to insulin) often overexpress PDHK4.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for PDHK4 in diverse human cancers of 329, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 24751 diverse cancer specimens. This rate is very similar (+ 9% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.54 % in 864 skin cancers tested; 0.44 % in 603 endometrium cancers tested; 0.29 % in 1270 large intestine cancers tested; 0.16 % in 1512 liver cancers tested; 0.14 % in 710 oesophagus cancers tested; 0.12 % in 589 stomach cancers tested; 0.09 % in 273 cervix cancers tested; 0.07 % in 1316 breast cancers tested; 0.06 % in 1276 kidney cancers tested; 0.05 % in 2009 haematopoietic and lymphoid cancers tested; 0.05 % in 1459 pancreas cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R124Q (3); R124* (3).
Comments:
Only 2 deletions, and no insertions or complex mutations are noted on the COSMIC website.

