Nomenclature
Short Name:
Slob
Full Name:
PX domain-containing protein kinase-like protein
Alias:
- Modulator of Na,K-ATPase
- MONaKA
- PXK
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
Slob
SubFamily:
NA
Structure
Mol. Mass (Da):
64,950
# Amino Acids:
578
# mRNA Isoforms:
7
mRNA Isoforms:
64,950 Da (578 AA; Q7Z7A4); 62,891 Da (561 AA; Q7Z7A4-4); 60,956 Da (545 AA; Q7Z7A4-6); 58,732 Da (515 AA; Q7Z7A4-2); 55,762 Da (495 AA; Q7Z7A4-7); 49,491 Da (441 AA; Q7Z7A4-5); 38,875 Da (352 AA; Q7Z7A4-3)
4D Structure:
NA
1D Structure:
Subfamily Alignment
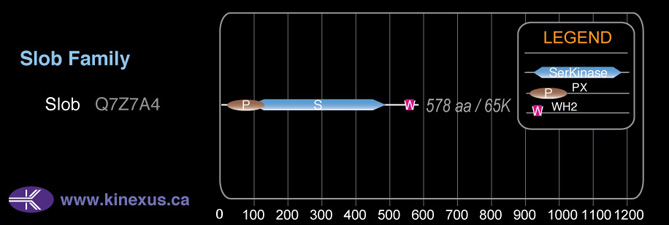
Domain Distribution:
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Methylated:
K568, K576.
Serine phosphorylated:
S57, S448, S452, S467, S470, S488, S507.
Threonine phosphorylated:
T444, T563.
Tyrosine phosphorylated:
Y56, Y163.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 64
64
2527
21
1669
 20
20
775
8
1303
 -
-
-
-
-
 15
15
584
90
977
 22
22
880
27
474
 4
4
161
34
208
 1.1
1.1
43
33
15
 50
50
1976
19
3867
 0.7
0.7
27
3
1
 7
7
273
37
734
 12
12
462
13
1036
 100
100
3951
42
6031
 33
33
1286
2
138
 33
33
1319
5
1995
 8
8
304
13
670
 9
9
353
17
822
 6
6
249
239
2252
 15
15
576
5
763
 18
18
724
23
2171
 31
31
1221
79
829
 24
24
963
13
1763
 2
2
91
11
104
 -
-
-
-
-
 52
52
2071
7
2128
 9
9
373
13
726
 23
23
898
60
826
 24
24
966
5
1275
 28
28
1107
5
1507
 24
24
938
5
1209
 1.3
1.3
51
14
42
 25
25
973
12
78
 4
4
147
23
247
 2
2
77
75
86
 22
22
870
78
633
 5
5
208
48
144
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.7
99.7
99.7
100 77
77
78.5
- -
-
-
97 -
-
-
- 84.6
84.6
85.3
94 -
-
-
- 92.3
92.3
95.5
92 92.4
92.4
95.7
92 -
-
-
- 25.9
25.9
27.7
- 83.4
83.4
90.5
85 80.6
80.6
89.6
81.5 73.4
73.4
85.5
73 -
-
-
- 35.9
35.9
54
44 42.9
42.9
61.1
- -
-
-
- 35.8
35.8
50.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
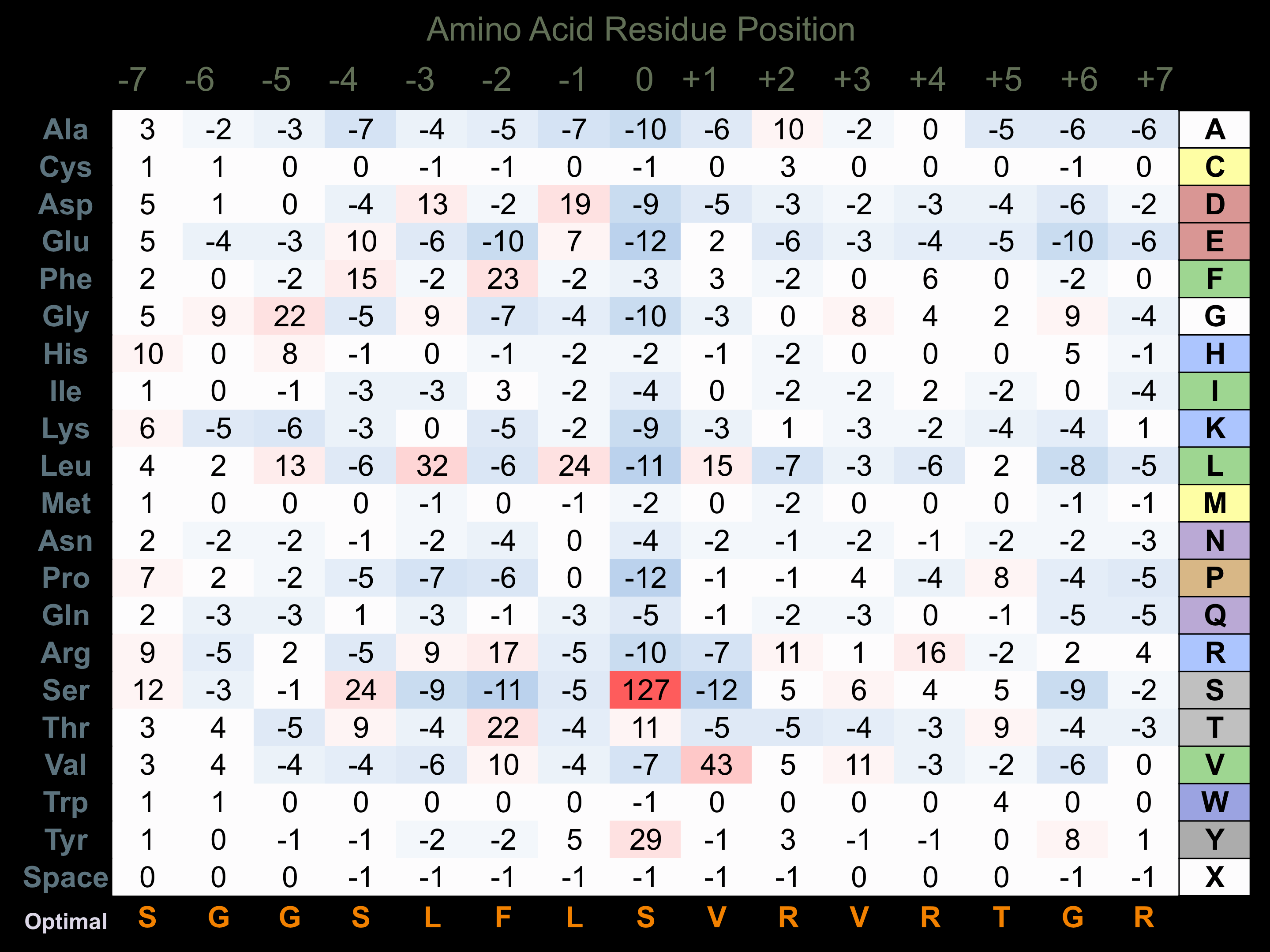
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Immune disorders
Specific Diseases (Non-cancerous):
Systemic lupus erythematosus (SLE); Systemic lupus erythematosus (SLE) susceptibility
Comments:
Systemic lupus erythematosus (SLE) is an autoimmune disorder characterized by widespread inflammation and tissue destruction resulting from the production of antibodies that target the body's own tissues (auto-antibodies). SLE is idiopathic, however links to a variety of genetic, environmental, and hormonal factors have been proposed. Symptoms of SLE include fatigue, pain or swelling in joints, skin inflammation, and fever which may present in periods of illness and remission. The Slob gene has been suggested as a suceptibility gene for SLE as a result of genome-wide association studies (GWAS). It was reported that the rs6445975 polymorphism of the Slob gene was significantly associated with the development of SLE is Caucasian populations. However, similar associations between the gene and disease have not been observed for either Chinese or South Korea populations.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24434 diverse cancer specimens. This rate is only -12 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.34 % in 555 stomach cancers tested; 0.31 % in 1229 large intestine cancers tested; 0.23 % in 382 soft tissue cancers tested; 0.2 % in 603 endometrium cancers tested; 0.19 % in 548 urinary tract cancers tested; 0.16 % in 864 skin cancers tested; 0.15 % in 1512 liver cancers tested; 0.06 % in 273 cervix cancers tested; 0.05 % in 1437 pancreas cancers tested; 0.04 % in 1608 lung cancers tested; 0.03 % in 558 thyroid cancers tested; 0.03 % in 1983 haematopoietic and lymphoid cancers tested; 0.03 % in 1289 breast cancers tested; 0.02 % in 881 prostate cancers tested; 0.02 % in 807 ovary cancers tested; 0.01 % in 2030 central nervous system cancers tested; 0.01 % in 1253 kidney cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: A369V (3); P572H (3).
Comments:
Only 2 deletions, 3 insertions, and 1 complex mutation are noted on the COSMIC website.

