Nomenclature
Short Name:
AlphaK2
Full Name:
Alpha-protein kinase 2
Alias:
- Alpha-kinase 2
- ALPK2
- EC 2.7.11.-
- HAK
- Heart alpha-kinase
Classification
Type:
Protein-serine/threonine kinase
Group:
Atypical
Family:
Alpha
SubFamily:
NA
Structure
Mol. Mass (Da):
237,013
# Amino Acids:
2170
# mRNA Isoforms:
1
mRNA Isoforms:
237,013 Da (2170 AA; Q86TB3)
4D Structure:
NA
1D Structure:
Subfamily Alignment
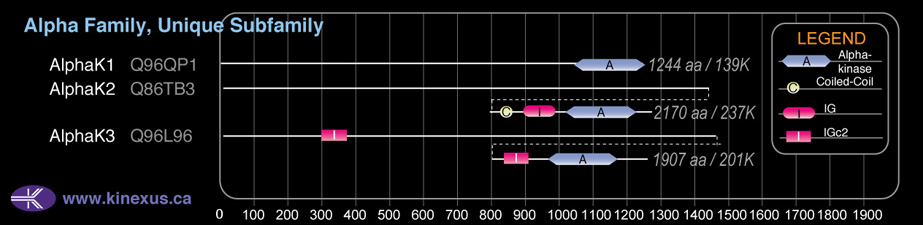
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 1482 | 1509 | Coiled-coil |
| 1786 | 1874 | IG |
| 1901 | 2133 | Alpha_kinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K528.
Serine phosphorylated:
S621, S623, S1602.
Threonine phosphorylated:
T617, T1868, T2089, T2095.
Tyrosine phosphorylated:
Y1975.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 36
36
1217
15
1163
 0.1
0.1
3
5
1
 1.3
1.3
45
8
33
 15
15
522
64
2391
 12
12
412
22
296
 0.2
0.2
7
18
7
 0.3
0.3
9
23
7
 25
25
836
19
815
 0.1
0.1
3
3
1
 10
10
333
63
301
 3
3
90
15
65
 13
13
444
27
486
 2
2
71
8
50
 0.3
0.3
9
3
3
 0.6
0.6
20
14
19
 0.2
0.2
7
10
6
 1.5
1.5
50
144
44
 0.9
0.9
30
11
36
 12
12
415
47
403
 10
10
349
59
213
 0.8
0.8
26
12
28
 0.8
0.8
27
15
23
 0.9
0.9
30
8
28
 0.6
0.6
21
11
18
 4
4
134
15
145
 44
44
1472
46
4622
 4
4
119
11
77
 0.6
0.6
20
8
18
 0.4
0.4
15
11
19
 6
6
210
28
137
 29
29
993
12
112
 100
100
3369
16
6873
 0.1
0.1
2
24
0
 20
20
672
52
578
 14
14
459
35
614
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 93.6
93.6
94.4
95.5 78
78
81
- -
-
-
61 -
-
-
- 62
62
71.9
64 -
-
-
- 51
51
61.1
57 36.4
36.4
47.6
56 -
-
-
- 34.3
34.3
47.8
- 30.1
30.1
46.3
- -
-
-
- 22.6
22.6
40.7
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Disease Linkage
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in human Classical Hodgkin lymphomas (%CFC= +232, p<0.008). The COSMIC website notes an up-regulated expression score for AlphaK2 in diverse human cancers of 324, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 24812 diverse cancer specimens. This rate is only -21 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.21 % in 864 skin cancers tested; 0.11 % in 1270 large intestine cancers tested; 0.08 % in 603 endometrium cancers tested; 0.06 % in 1634 lung cancers tested; 0.05 % in 589 stomach cancers tested; 0.05 % in 273 cervix cancers tested.
Frequency of Mutated Sites:
None > 6 in 19,715 cancer specimens
Comments:
Ten deletions mutations are all located at F750.

