Nomenclature
Short Name:
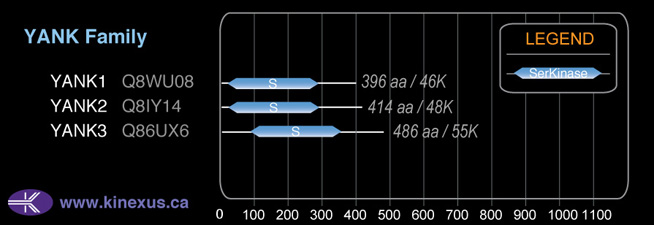
YANK3
Full Name:
Serine/threonine-protein kinase 32C
Alias:
- EC 2.7.11.1
- PKE
- ST32C
- STK32C
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
YANK
SubFamily:
NA
Structure
Mol. Mass (Da):
54,994
# Amino Acids:
486
# mRNA Isoforms:
2
mRNA Isoforms:
54,994 Da (486 AA; Q86UX6); 42,425 Da (369 AA; Q86UX6-2)
4D Structure:
NA
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 93 | 353 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S10, S11, S15, S18.
Tyrosine phosphorylated:
Y119.
Ubiquitinated:
K218, K364.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 50
50
1390
15
901
 4
4
104
4
71
 -
-
-
-
-
 22
22
622
62
1632
 24
24
668
20
409
 0.8
0.8
21
18
12
 0.8
0.8
21
20
14
 20
20
562
9
1029
 -
-
-
-
-
 4
4
121
56
66
 4
4
107
8
79
 32
32
886
20
582
 5
5
131
2
39
 5
5
141
2
117
 8
8
219
8
113
 2
2
53
12
34
 5
5
133
193
91
 3
3
88
2
24
 5
5
136
38
84
 18
18
501
58
297
 8
8
234
8
144
 8
8
232
6
98
 -
-
-
-
-
 9
9
263
4
79
 9
9
254
8
109
 78
78
2157
39
3878
 2
2
63
2
5
 6
6
177
2
70
 6
6
156
2
33
 9
9
250
28
178
 30
30
830
12
97
 84
84
2329
18
3736
 0.2
0.2
6
24
4
 30
30
831
52
704
 100
100
2773
35
3997
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.3
99.3
99.5
99 56.7
56.7
69.1
- -
-
-
93 -
-
-
- 71.2
71.2
77.1
92 -
-
-
- 90.9
90.9
94
91 91.3
91.3
94.4
92 -
-
-
- 50.3
50.3
62.3
- 65.4
65.4
71.1
83 52.8
52.8
65.4
79 -
-
-
- -
-
-
- 25.7
25.7
45
37 40.3
40.3
54.1
- -
-
-
- 47
47
61.9
- -
-
-
- -
-
-
- -
-
-
- 30.8
30.8
48.1
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
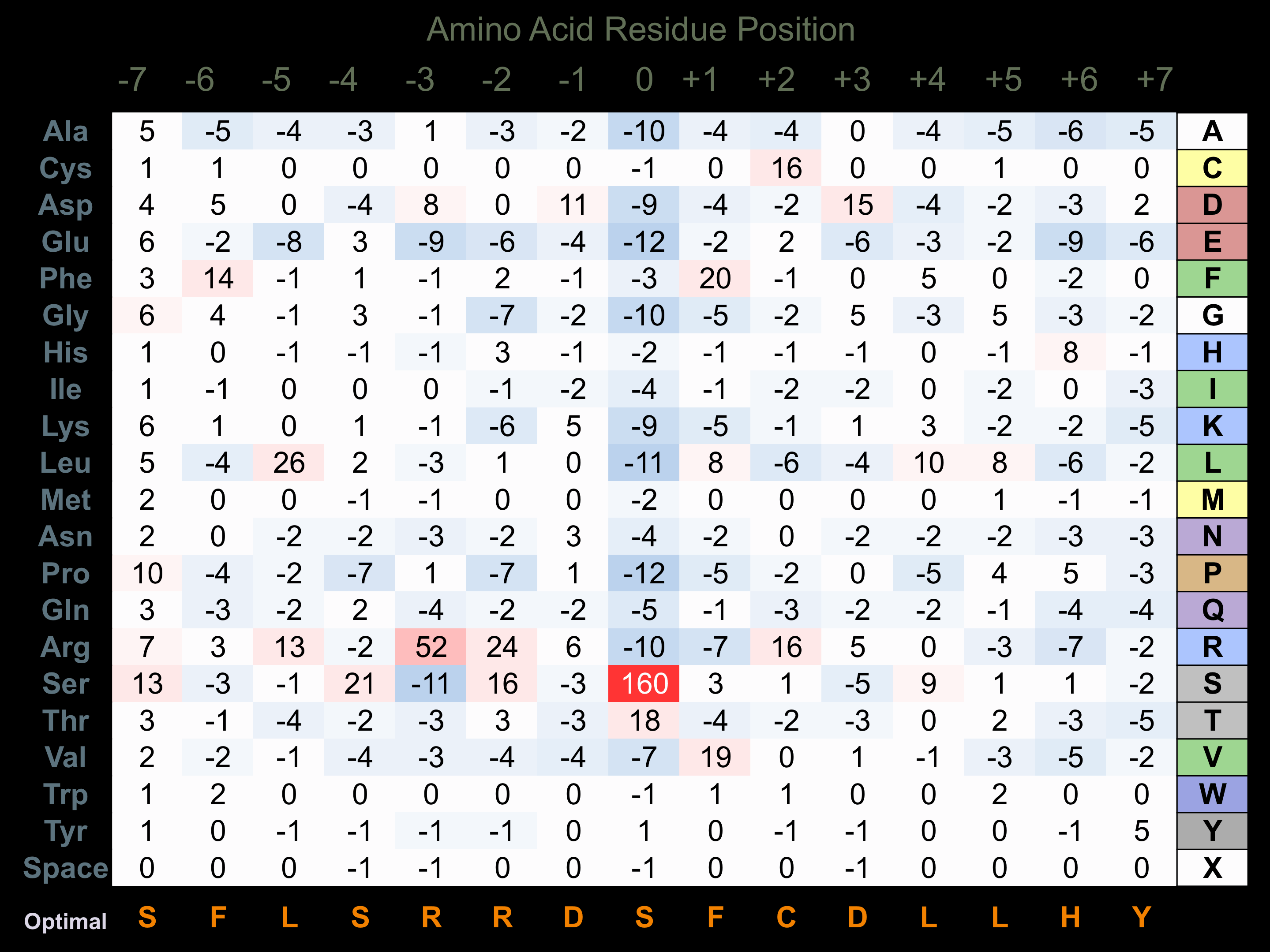
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Bone disorder
Specific Diseases (Non-cancerous):
Ellis-Van Creveld syndrome
Comments:
Ellis-Van Creveld syndrome, also known as chondroectodermal dysplasia, is an inherited bone disease characterized by very short statue (dwarfism) as a result of significantly impaired bone growth during development. Characteristic features of the disease include short forearms and lower legs, narrow chest, short ribs, presence of extra digits (polydactyly), malformed toenails and fingernails, and abnormal dentition. Approximately 60% of affected individuals have serious congenital heart defects, which can be life-threatening. Homozygous loss-of-function mutations in the EVC or EVC2 genes have been previously linked with the disease. Additionally, a 520-kb homozygous deletion mutation comprising the gene sequences for EVC, EVC2, C4of6, and YANK2 was reported in patients diagnosed with Ellis-Van Creveld syndrome and borderline intelligence. Furthermore, the phenotype of the affected patients indicated that the EVC-YANK2 deletion results in a mild degree of mental retardation and that deletion of the C4of6 and YANK2 gene results in mild mental deficit at most. Mutations in YANK2 have also been associated with isolated cases of cleft lip and cleft palate, whcih are two of the most common birth defects in human populations.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for YANK3 in diverse human cancers of 422, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 23 for this protein kinase in human cancers was 0.4-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 25414 diverse cancer specimens. This rate is only -18 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.22 % in 1299 large intestine cancers tested; 0.2 % in 1956 lung cancers tested; 0.17 % in 864 skin cancers tested; 0.17 % in 603 endometrium cancers tested; 0.11 % in 548 urinary tract cancers tested; 0.09 % in 942 upper aerodigestive tract cancers tested; 0.07 % in 589 stomach cancers tested; 0.05 % in 891 ovary cancers tested; 0.05 % in 441 autonomic ganglia cancers tested; 0.05 % in 382 soft tissue cancers tested; 0.05 % in 1529 breast cancers tested; 0.05 % in 1512 liver cancers tested; 0.04 % in 1467 pancreas cancers tested; 0.02 % in 939 prostate cancers tested; 0.02 % in 2009 haematopoietic and lymphoid cancers tested; 0.02 % in 1276 kidney cancers tested; 0.01 % in 2082 central nervous system cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: S392Y (4).
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.

