Nomenclature
Short Name:
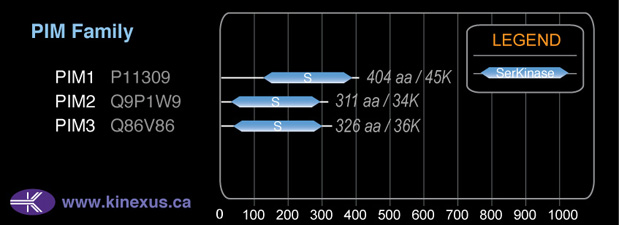
PIM3
Full Name:
Serine-threonine-protein kinase Pim-3
Alias:
- EC 2.7.11.1
- Pim-3 oncogene
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
PIM
SubFamily:
NA
Structure
Mol. Mass (Da):
35891
# Amino Acids:
326
# mRNA Isoforms:
1
mRNA Isoforms:
35,891 Da (326 AA; Q86V86)
4D Structure:
Interacts with BAD.
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 40 | 293 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S8, S81+, S192+, S211-.
Threonine phosphorylated:
T77, T86, T202+, T207-.
Ubiquitinated:
K30, K38, K69, K172, K186, K197.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 68
68
1993
6
1133
 6
6
178
8
156
 -
-
-
-
-
 15
15
441
40
409
 46
46
1343
13
804
 9
9
254
9
97
 4
4
116
13
62
 15
15
450
17
565
 0.6
0.6
18
3
5
 24
24
704
37
426
 12
12
342
13
560
 46
46
1326
19
837
 9
9
256
4
77
 6
6
170
7
199
 16
16
471
13
640
 11
11
333
9
261
 28
28
824
212
1787
 4
4
124
7
70
 19
19
563
32
381
 49
49
1438
35
866
 13
13
366
13
333
 19
19
561
9
486
 -
-
-
-
-
 14
14
401
11
350
 13
13
379
13
345
 46
46
1332
27
1146
 8
8
233
7
389
 19
19
560
7
623
 9
9
263
7
312
 16
16
465
14
152
 44
44
1276
12
31
 100
100
2913
15
4185
 21
21
606
12
269
 44
44
1276
26
755
 17
17
497
13
335
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 45.5
45.5
53.2
0 71.1
71.1
72.3
96 -
-
-
95 -
-
-
- 48.2
48.2
57
- -
-
-
- 95.7
95.7
96.9
96 95.7
95.7
96.9
96 -
-
-
- 50
50
62.4
- -
-
-
81 76.4
76.4
85.6
80 54.3
54.3
71.2
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | BAD - Q92934 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
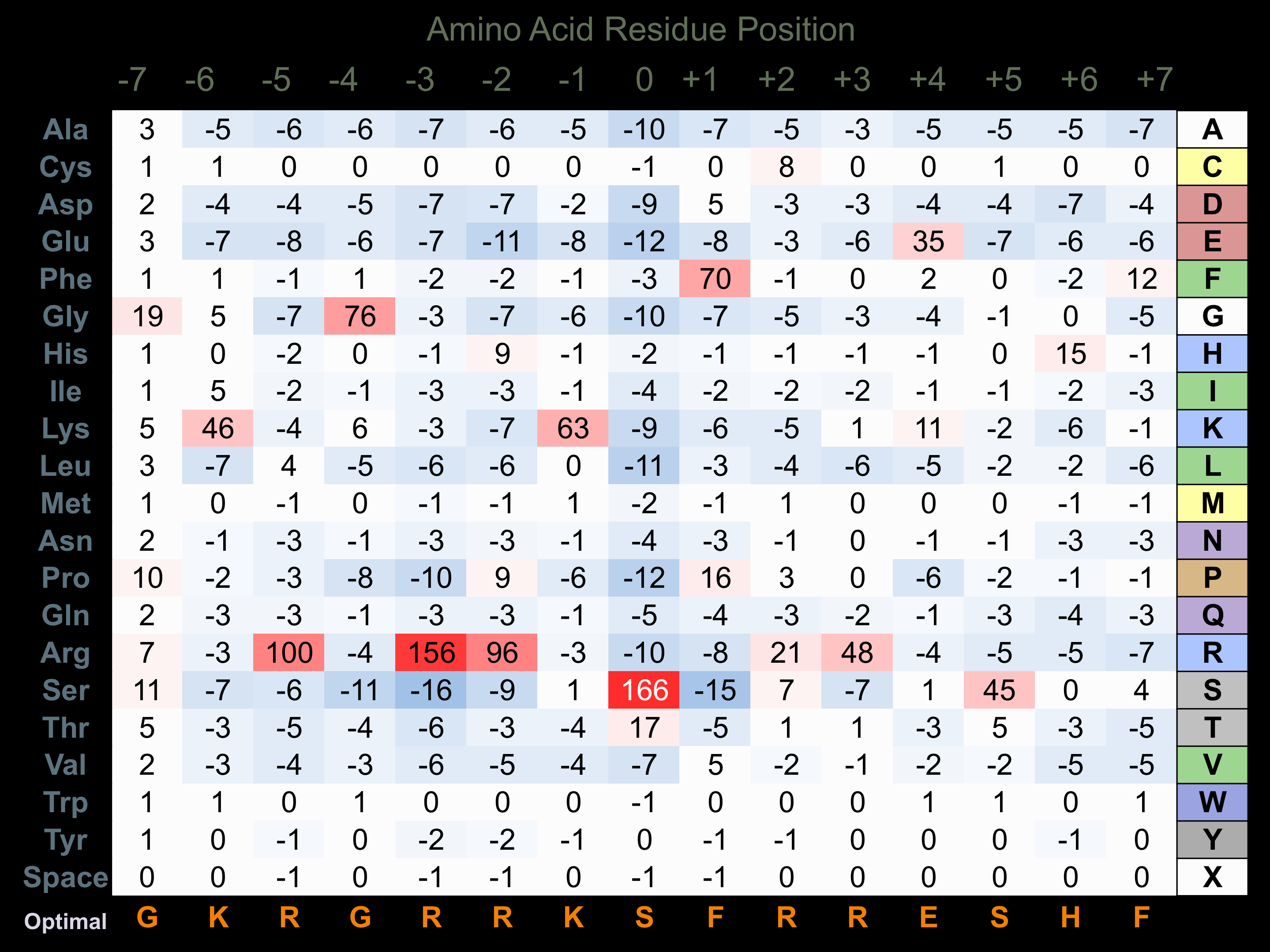
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Cancer, neurological disorders
Specific Diseases (Non-cancerous):
Cannabis dependence
Specific Cancer Types:
Pancreatic cancer
Comments:
Pim3 appears to be a tumour requiring protein (TRP), since it undergoes fewer mutations than the typical protein in cancer cells. The active form of the protein kinase normally acts to promote tumour cell proliferation. Pim3 promotes tumorigenesis, and is upregulated in human pancreatic cancer tissue and pancreatic cancer cell lines.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain oligodendrogliomas (%CFC= +273, p<0.0001); Breast epithelial carcinomas (%CFC= +96, p<0.054); Classical Hodgkin lymphomas (%CFC= +131, p<0.0006); Colon mucosal cell adenomas (%CFC= +49, p<0.0001); Colorectal adenocarcinomas (early onset) (%CFC= +50, p<0.064); and Large B-cell lymphomas (%CFC= +119, p<0.0001).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.04 % in 24433 diverse cancer specimens. This rate is -43 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
None >2 in 24433 cancer specimens
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.

