Nomenclature
Short Name:
SgK085
Full Name:
Myosin light chain kinase family member 4
Alias:
- LOC134927
- NP_001012418
- Sugen kinase 85
- MYLK4
- EC=2.7.11.1
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
MLCK
SubFamily:
NA
Structure
Mol. Mass (Da):
44,508
# Amino Acids:
388
# mRNA Isoforms:
2
mRNA Isoforms:
44,508 Da (388 AA; Q86YV6); 44,487 Da (388 AA; Q86YV6-2)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
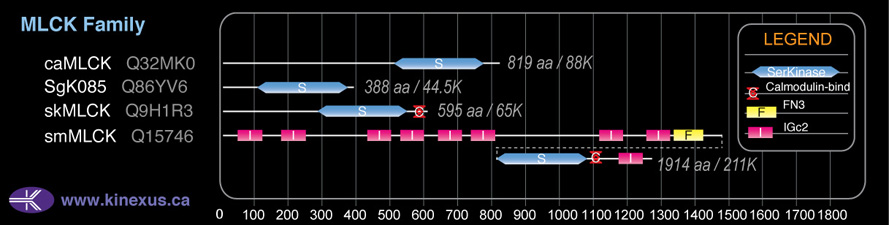
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 106 | 361 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1657
18
1293
 0.4
0.4
6
6
5
 -
-
-
-
-
 4
4
66
75
273
 35
35
572
24
256
 0.5
0.5
9
27
8
 0.4
0.4
6
33
5
 0.2
0.2
3
4
3
 0.2
0.2
3
3
1
 0.2
0.2
4
3
2
 0.4
0.4
6
3
4
 49
49
806
21
376
 -
-
-
-
-
 0.3
0.3
5
3
1
 1
1
19
3
24
 0.7
0.7
12
15
9
 0.4
0.4
7
15
6
 0.3
0.3
5
3
0
 0.5
0.5
8
3
3
 28
28
462
72
182
 0.2
0.2
3
3
1
 0.2
0.2
3
3
1
 -
-
-
-
-
 0.2
0.2
4
3
2
 0.4
0.4
7
3
5
 44
44
733
53
1230
 0.2
0.2
3
3
1
 0.2
0.2
4
3
1
 0.3
0.3
5
3
1
 -
-
-
-
-
 50
50
829
12
94
 5
5
80
15
17
 0.1
0.1
2
36
1
 48
48
801
78
725
 9
9
157
44
184
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.2
99.2
100
99 96.9
96.9
98.1
98 -
-
-
85 -
-
-
- 83.4
83.4
87.6
89 -
-
-
- 84.7
84.7
90.7
87 37.5
37.5
48.3
89 -
-
-
- 55.8
55.8
66.4
- -
-
-
83 -
-
-
77 -
-
-
72 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | ALB - P02768 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
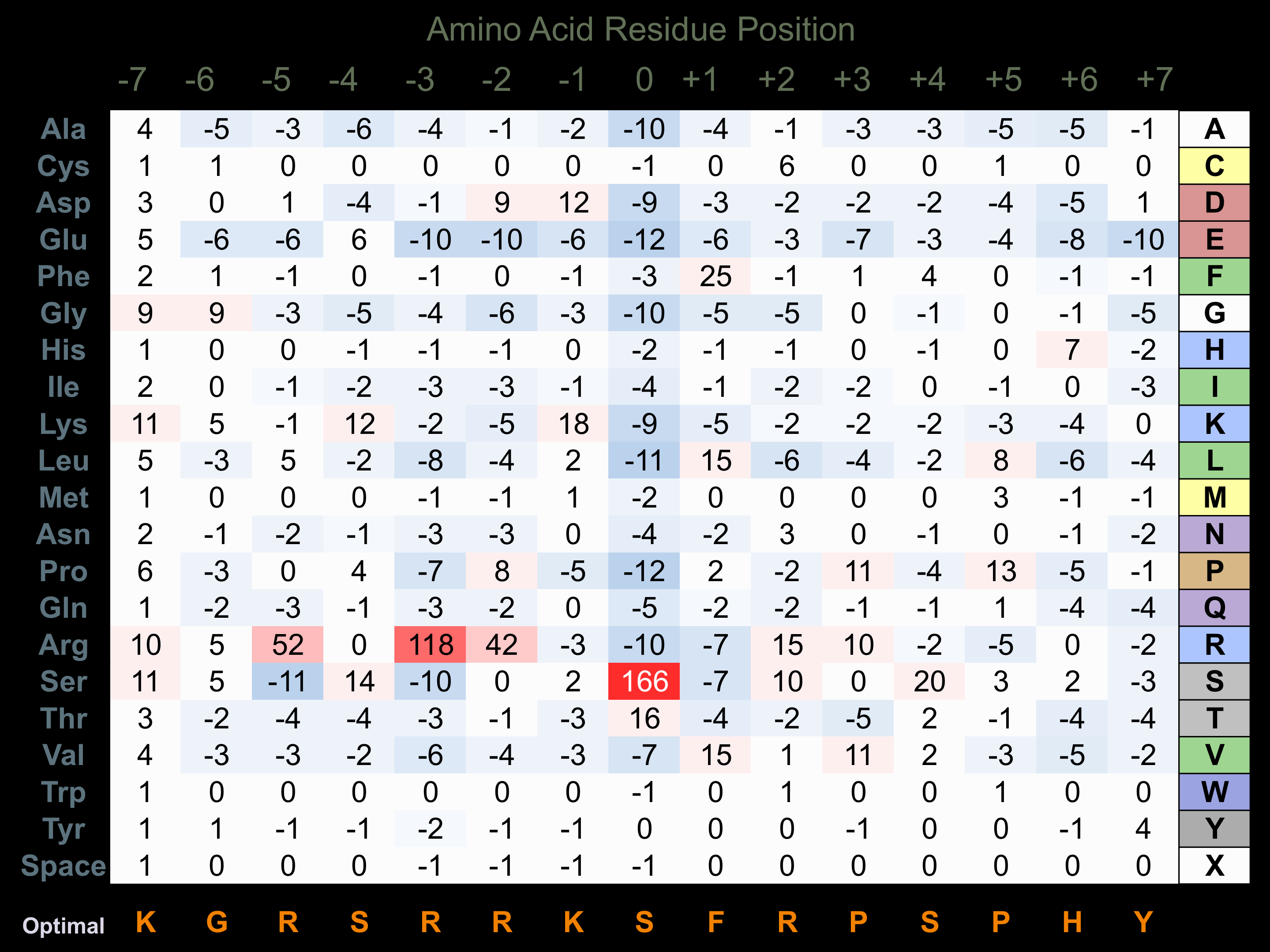
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Sunitinib | Kd = 15 nM | 5329102 | 535 | 18183025 |
| Lestaurtinib | Kd = 29 nM | 126565 | 22037378 | |
| SU14813 | Kd = 32 nM | 10138259 | 1721885 | 18183025 |
| Dovitinib | Kd = 80 nM | 57336746 | 18183025 | |
| TG101348 | Kd = 130 nM | 16722836 | 1287853 | 22037378 |
| KW2449 | Kd = 170 nM | 11427553 | 1908397 | 22037378 |
| Enzastaurin | Kd = 230 nM | 176167 | 300138 | 22037378 |
| Nintedanib | Kd = 440 nM | 9809715 | 502835 | 22037378 |
| Staurosporine | Kd = 470 nM | 5279 | 18183025 | |
| N-Benzoylstaurosporine | Kd = 640 nM | 56603681 | 608533 | 18183025 |
| BMS-345541 | Kd = 700 nM | 9813758 | 249697 | 22037378 |
| GDC-0941 | Kd = 780 nM | 17755052 | 521851 | 22037378 |
| R406 | Kd = 810 nM | 11984591 | 22037378 | |
| JNJ-28312141 | Kd = 870 nM | 22037378 | ||
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| A674563 | Kd = 1.5 µM | 11314340 | 379218 | 22037378 |
| Tozasertib | Kd = 2.2 µM | 5494449 | 572878 | 18183025 |
| AC1NS7CD | Kd = 2.4 µM | 5329665 | 295136 | 22037378 |
| SNS032 | Kd = 2.4 µM | 3025986 | 296468 | 22037378 |
| PLX4720 | Kd = 3.4 µM | 24180719 | 1230020 | 22037378 |
Disease Linkage
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for SgK085 in diverse human cancers of 391, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 10 for this protein kinase in human cancers was 0.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.11 % in 24682 diverse cancer specimens. This rate is a modest 1.48-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 1.72 % in 15 pituitary cancers tested; 0.61 % in 1270 large intestine cancers tested; 0.57 % in 864 skin cancers tested; 0.34 % in 603 endometrium cancers tested; 0.22 % in 589 stomach cancers tested; 0.21 % in 1634 lung cancers tested; 0.15 % in 710 oesophagus cancers tested; 0.1 % in 807 ovary cancers tested; 0.09 % in 273 cervix cancers tested; 0.09 % in 1512 liver cancers tested; 0.09 % in 1421 breast cancers tested; 0.06 % in 881 prostate cancers tested; 0.06 % in 1276 kidney cancers tested; 0.05 % in 548 urinary tract cancers tested; 0.04 % in 2074 central nervous system cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: E30G(4).
Comments:
Only 1 deletion, 2 insertions and no complex mutations are noted on the COSMIC website.

