Nomenclature
Short Name:
SgK307
Full Name:
Testis expressed protein 14
Alias:
- Sugen kinase 307
- TEX14
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
NKF5
SubFamily:
NA
Structure
Mol. Mass (Da):
167,901
# Amino Acids:
1497
# mRNA Isoforms:
3
mRNA Isoforms:
167,901 Da (1497 AA; Q8IWB6); 167,140 Da (1491 AA; Q8IWB6-2); 162,669 Da (1451 AA; Q8IWB6-3)
4D Structure:
NA
1D Structure:
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 27 | 54 | ANK |
| 55 | 87 | ANK |
| 88 | 120 | ANK |
| 244 | 518 | Pkinase |
| 394 | 470 | Pkinase |
| 650 | 670 | Coiled-coil |
| 741 | 768 | Coiled-coil |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K511, K515, K877, K925, K935.
Serine phosphorylated:
S284, S437, S915, S1280, S1305, S1384, S1385, S1493, S1496.
Threonine phosphorylated:
T618, T727, T728, T1277.
Tyrosine phosphorylated:
Y36, Y206, Y508, Y1187.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 12
12
340
16
672
 0.3
0.3
10
8
6
 12
12
358
21
1106
 23
23
657
64
1844
 10
10
296
14
293
 0.2
0.2
7
37
21
 0.2
0.2
7
15
7
 30
30
879
32
1892
 8
8
219
10
219
 3
3
80
50
273
 7
7
189
32
520
 19
19
552
83
548
 5
5
150
32
460
 0.3
0.3
10
6
3
 4
4
129
29
386
 0.1
0.1
3
7
3
 7
7
201
33
682
 5
5
140
26
322
 2
2
47
57
116
 8
8
234
56
283
 5
5
133
28
315
 5
5
130
24
376
 7
7
214
22
598
 43
43
1252
26
1537
 6
6
175
28
474
 13
13
381
45
664
 3
3
93
31
321
 5
5
157
26
430
 5
5
133
26
314
 1.1
1.1
31
14
12
 14
14
401
30
205
 100
100
2886
21
6378
 0.1
0.1
3
17
1
 35
35
1004
26
844
 8
8
231
31
226
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 98.7
98.7
99
99 -
-
-
92.5 -
-
-
77 -
-
-
- 76.3
76.3
83.3
78.5 -
-
-
- 63.3
63.3
74.8
68.5 64.5
64.5
75
70 -
-
-
- 37.8
37.8
52.6
- 36.6
36.6
50
44 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
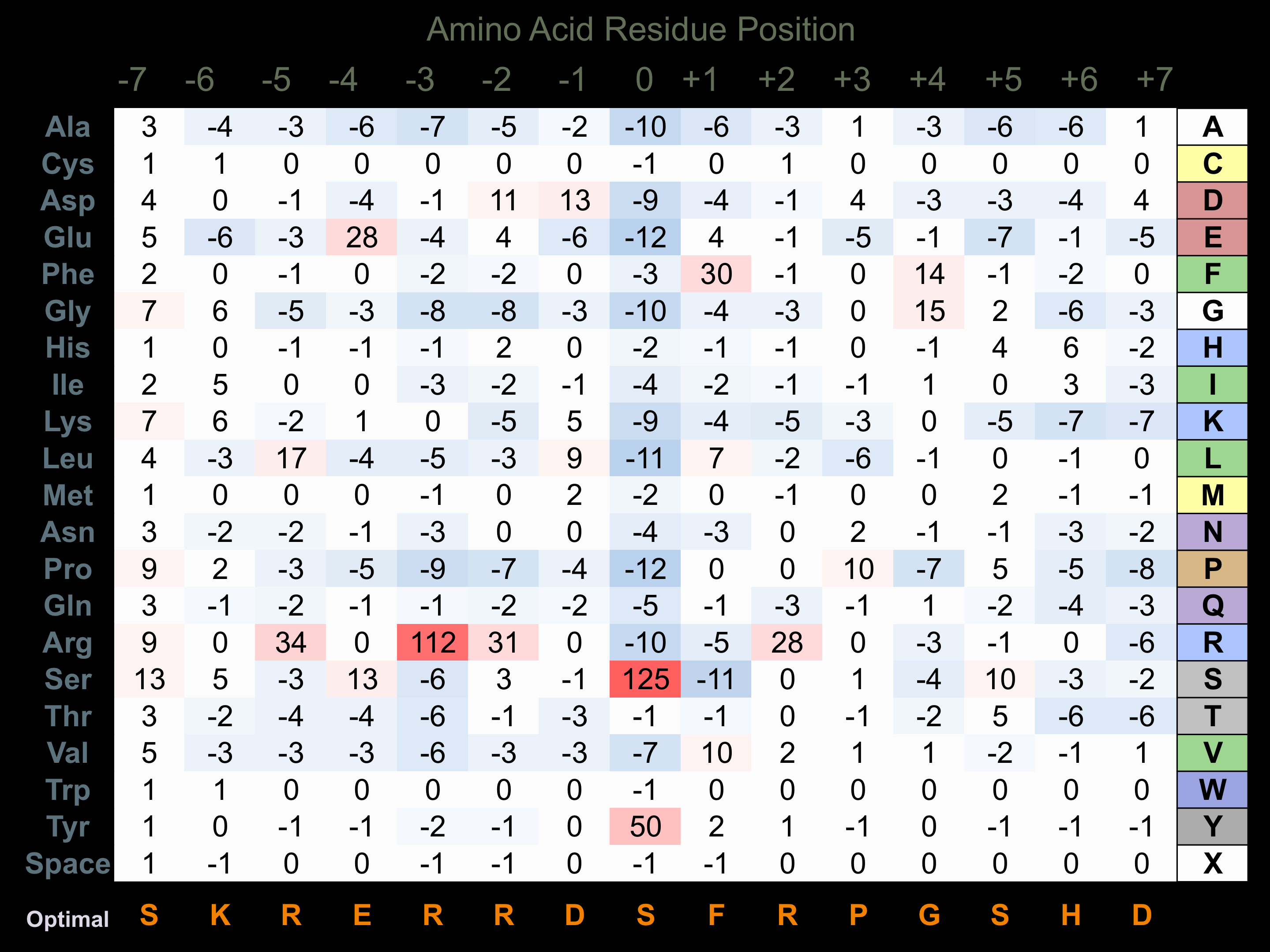
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= -48, p<0.0002); and Brain oligodendrogliomas (%CFC= -59, p<0.0001).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24449 diverse cancer specimens. This rate is only -1 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.37 % in 864 skin cancers tested; 0.34 % in 1230 large intestine cancers tested; 0.2 % in 603 endometrium cancers tested; 0.2 % in 589 stomach cancers tested; 0.17 % in 238 bone cancers tested; 0.16 % in 548 urinary tract cancers tested; 0.14 % in 1608 lung cancers tested; 0.1 % in 273 cervix cancers tested; 0.09 % in 710 oesophagus cancers tested; 0.06 % in 1512 liver cancers tested; 0.04 % in 942 upper aerodigestive tract cancers tested; 0.04 % in 1289 breast cancers tested; 0.04 % in 1253 kidney cancers tested; 0.03 % in 881 prostate cancers tested; 0.03 % in 807 ovary cancers tested; 0.03 % in 1437 pancreas cancers tested; 0.02 % in 558 thyroid cancers tested; 0.02 % in 441 autonomic ganglia cancers tested; 0.01 % in 2030 central nervous system cancers tested.
Frequency of Mutated Sites:
None > 5 in 20,484 cancer specimens
Comments:
Only 4 deletions, 4 insertions, and no complex mutations are noted on the COSMIC website.

