Nomenclature
Short Name:
CCRK
Full Name:
Cell cycle-related kinase
Alias:
- Cyclin-kinase activating kinase p42
- CDK-activating kinase p42
- CAK-kinase p42
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
CDK
SubFamily:
NA
Structure
Mol. Mass (Da):
38,695
# Amino Acids:
346
# mRNA Isoforms:
5
mRNA Isoforms:
38,695 Da (346 AA; Q8IZL9); 37,592 Da (338 AA; Q8IZL9-4); 36,113 Da (325 AA; Q8IZL9-5); 30,571 Da (275 AA; Q8IZL9-3); 27,100 Da (243 AA; Q8IZL9-2)
4D Structure:
Monomer. Interacts with BROMI
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 4 | 288 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Threonine phosphorylated:
T161.
Tyrosine phosphorylated:
Y4.
Ubiquitinated:
K33, K53, K129, K239.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 19
19
1013
16
1338
 2
2
91
10
42
 2
2
128
7
63
 6
6
334
54
487
 11
11
611
14
570
 0.4
0.4
19
37
10
 5
5
289
19
495
 8
8
439
23
667
 4
4
206
10
191
 2
2
105
44
66
 1
1
55
20
59
 10
10
545
103
626
 1
1
67
18
62
 0.8
0.8
44
9
23
 2
2
88
17
96
 2
2
113
8
35
 1
1
66
167
425
 1
1
63
12
50
 0.9
0.9
49
49
30
 5
5
295
56
343
 2
2
85
17
52
 1
1
55
18
59
 2
2
96
8
76
 4
4
232
14
104
 1
1
59
17
69
 11
11
591
33
752
 0.8
0.8
45
21
35
 2
2
94
13
51
 2
2
94
13
65
 3
3
143
14
75
 11
11
601
18
407
 100
100
5418
21
10368
 5
5
247
46
503
 16
16
868
26
734
 2
2
105
22
78
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
95 57.5
57.5
60.6
- -
-
-
92.5 -
-
-
- 93.3
93.3
97.1
93 -
-
-
- 94.2
94.2
97.1
94 93.6
93.6
96.5
94 -
-
-
- 84.9
84.9
93
- -
-
-
- 39.4
39.4
55.1
- 75.4
75.4
86.9
74 -
-
-
- -
-
-
44 41.3
41.3
60.9
- -
-
-
- 69
69
82.9
- 40.1
40.1
54.3
- 38.1
38.1
55.2
- -
-
-
- 40.1
40.1
54.9
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CDK2 - P24941 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
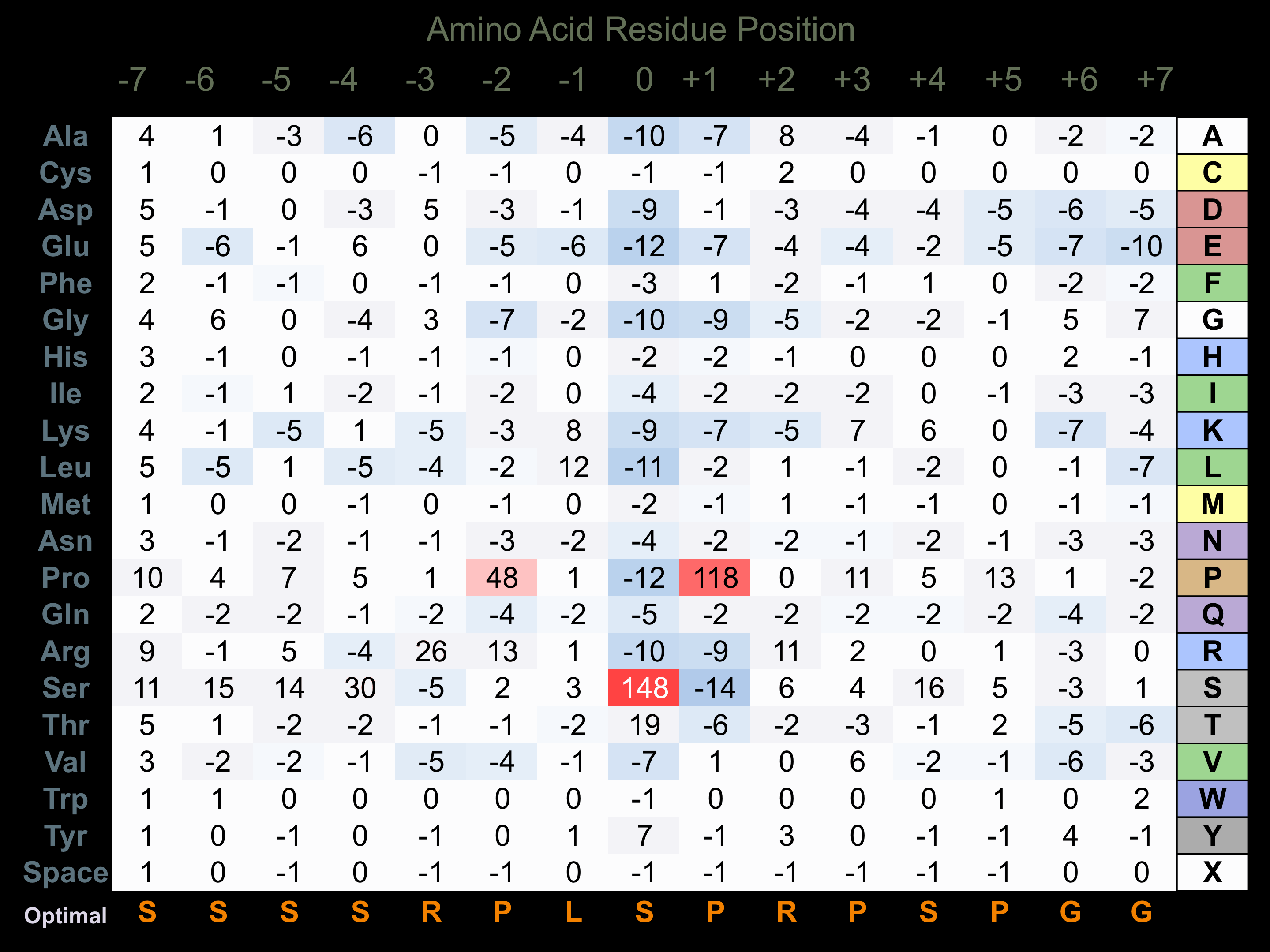
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Mantle cell lymphomas
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for CCRK in diverse human cancers of 339, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 4 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 24433 diverse cancer specimens. This rate is very similar (+ 3% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: W228C (4).
Comments:
Only 5 deletions, 1 insertion and no complex mutations are noted on the COSMIC website.

