Nomenclature
Short Name:
ADCK4
Full Name:
AarF domain containing kinase 4
Alias:
- FLJ32632
Classification
Type:
Protein-serine/threonine kinase
Group:
Atypical
Family:
ABC1
SubFamily:
ABC1-A
Structure
Mol. Mass (Da):
60,069
# Amino Acids:
544
# mRNA Isoforms:
2
mRNA Isoforms:
60,069 Da (544 AA; Q96D53); 55,853 Da (503 AA; Q96D53-2)
4D Structure:
NA
1D Structure:
Subfamily Alignment
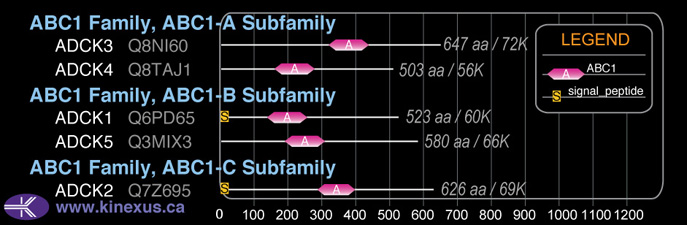
Domain Distribution:
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S79.
Ubiquitinated:
K424.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1024
25
1166
 57
57
581
16
1302
 44
44
455
4
790
 26
26
263
77
519
 57
57
582
26
584
 4
4
37
50
111
 2
2
24
28
21
 88
88
904
29
1685
 25
25
254
13
280
 19
19
196
62
747
 26
26
267
21
483
 58
58
589
90
504
 15
15
151
21
326
 24
24
248
13
513
 40
40
413
19
833
 20
20
203
16
415
 13
13
138
29
274
 17
17
173
18
262
 18
18
180
67
555
 38
38
394
91
431
 34
34
348
18
623
 12
12
122
16
313
 18
18
188
7
350
 67
67
681
22
1183
 27
27
274
18
517
 86
86
878
53
1526
 16
16
162
26
350
 28
28
283
18
472
 25
25
252
18
372
 23
23
234
28
250
 84
84
864
18
622
 32
32
324
30
362
 23
23
239
92
472
 74
74
756
52
641
 7
7
76
35
44
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 81.8
81.8
84
99.5 96.5
96.5
97.4
96.5 -
-
-
90.5 -
-
-
- 81.4
81.4
84.9
90 -
-
-
- 85.2
85.2
89.7
88 82.7
82.7
88.5
86 -
-
-
- 70.7
70.7
78.4
- 50.6
50.6
63.4
- 58.3
58.3
70.7
67 50.6
50.6
63.9
- -
-
-
- -
-
-
53 44.6
44.6
62.3
- 34.6
34.6
47.8
- 46.9
46.9
60.5
- -
-
-
- -
-
-
- -
-
-
45 -
-
-
45 37.5
37.5
53.6
41 -
-
-
47
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
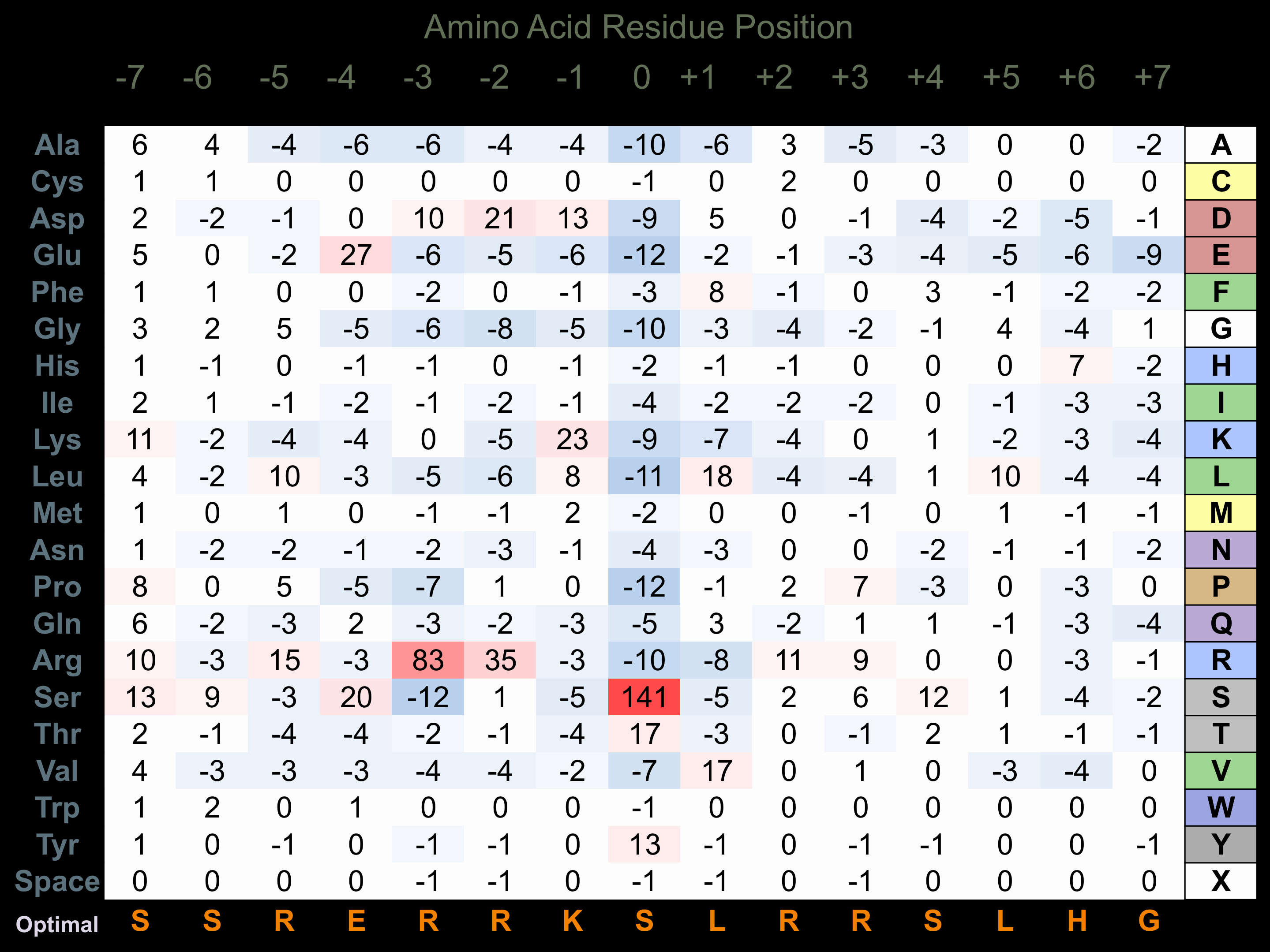
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| PD173955 | Kd = 27 nM | 447077 | 386051 | 22037378 |
| R406 | Kd = 64 nM | 11984591 | 22037378 | |
| NVP-TAE684 | Kd = 92 nM | 16038120 | 509032 | 22037378 |
| CHEMBL249097 | Kd = 129 nM | 25138012 | 249097 | 19035792 |
| TG101348 | Kd = 300 nM | 16722836 | 1287853 | 22037378 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| Crizotinib | Kd = 1.1 µM | 11626560 | 601719 | 22037378 |
| TG100115 | Kd = 1.4 µM | 10427712 | 230011 | 22037378 |
| Vandetanib | Kd = 1.7 µM | 3081361 | 24828 | 22037378 |
| Erlotinib | Kd = 2.5 µM | 176870 | 553 | 18183025 |
| SureCN7018367 | Kd < 2.5 µM | 18792927 | 450519 | 19035792 |
| AC1NS7CD | Kd = 2.9 µM | 5329665 | 295136 | 22037378 |
| SNS032 | Kd = 2.9 µM | 3025986 | 296468 | 22037378 |
| SB203580 | Kd = 3.1 µM | 176155 | 10 | 18183025 |
| Canertinib | Kd = 3.4 µM | 156414 | 31965 | 18183025 |
| PP242 | Kd = 3.5 µM | 25243800 | 22037378 | |
| BMS-690514 | Kd > 4 µM | 11349170 | 21531814 | |
| SB202190 | Kd = 4.9 µM | 5353940 | 278041 | 18183025 |
Disease Linkage
General Disease Association:
Coenzyme Q deficiency
Specific Diseases (Non-cancerous):
Steroid-resistant Nephrotic Syndrome Type 9 (NPHS9); Familial Idiopathic Steroid-Resistant Nephrotic Syndrome with Focal Segmental Hyalinosis
Comments:
Loss of ADCK4 function was linked to NPHS9. This included the following mutations: R178W; D286G; R320W; R343W and E483Stop as well as insertions and deletions.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in Ovary adenocarcinomas (%CFC= +95, p<0.043). The COSMIC website notes an up-regulated expression score for ADCK4 in diverse human cancers of 569, which is 1.2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 67 for this protein kinase in human cancers was 1.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 24712 diverse cancer specimens. This rate is only -16 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.34 % in 864 skin cancers tested; 0.3 % in 1270 large intestine cancers tested; 0.22 % in 589 stomach cancers tested; 0.21 % in 603 endometrium cancers tested.
Frequency of Mutated Sites:
None >3 in 19629 cancer specimens
Comments:
Only 3 deletions, no insertions or complex mutations are noted on the COSMIC website.

