Nomenclature
Short Name:
PBK
Full Name:
T-lymphokine-activated killer cell-originated protein kinase
Alias:
- CT84
- TOPK
- TPOK
- EC 2.7.12.2
- FLJ14385
- NORI3
- SPK
- T-LAK cell-originated protein kinase
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
TOPK
SubFamily:
NA
Structure
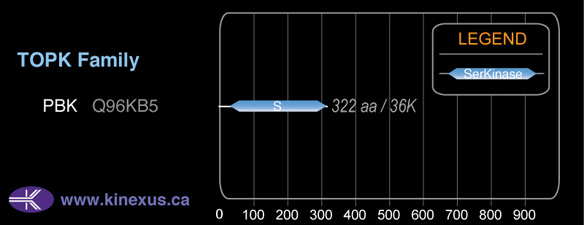
Mol. Mass (Da):
36,085
# Amino Acids:
322
# mRNA Isoforms:
2
mRNA Isoforms:
37,211 Da (333 AA; Q96KB5-2); 36,085 Da (322 AA; Q96KB5)
4D Structure:
Interacts with DLG1 and TP53.
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 29 | 322 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
M1, K12, K16, K17, K64.
Serine phosphorylated:
S14, S19, S23, S32, S57, S59, S76+, S91, S310.
Threonine phosphorylated:
T9+, T24, T26, T198+, T260.
Tyrosine phosphorylated:
Y47, Y74, Y78, Y100.
Ubiquitinated:
K50, K90, K132, K169.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 44
44
3494
16
7986
 0.1
0.1
9
8
13
 0.8
0.8
67
5
54
 24
24
1885
52
4304
 4
4
317
14
325
 1
1
76
37
50
 0.8
0.8
60
15
27
 100
100
7933
25
12958
 6
6
437
10
437
 0.2
0.2
15
37
12
 0.3
0.3
26
16
23
 6
6
504
67
428
 0.4
0.4
31
16
31
 0.2
0.2
15
6
7
 0.7
0.7
55
13
97
 0.1
0.1
9
7
6
 1
1
81
80
521
 0.3
0.3
25
10
32
 0.3
0.3
21
41
13
 5
5
408
56
470
 0.2
0.2
15
12
15
 0.3
0.3
20
13
17
 1.2
1.2
97
6
92
 8
8
659
10
534
 3
3
222
12
232
 74
74
5855
32
11876
 1
1
76
19
44
 0.3
0.3
24
10
19
 0.6
0.6
49
10
58
 0.6
0.6
51
14
28
 4
4
310
18
223
 96
96
7653
21
13130
 14
14
1087
53
1468
 9
9
688
26
596
 24
24
1888
22
2256
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.1
99.1
99.7
99 96.6
96.6
98.8
97 -
-
-
89 -
-
-
- 93.5
93.5
96.9
94 -
-
-
- 85.5
85.5
92.4
88 86.3
86.3
92
91 -
-
-
- 83.2
83.2
89.8
- 73.6
73.6
85.4
75 70.2
70.2
85.2
- 64
64
77.3
67 -
-
-
- 35.9
35.9
51.3
46 41.4
41.4
58.9
- -
-
-
- 46.5
46.5
64
- -
-
-
- 26.7
26.7
45.6
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CDC2 - P06493 |
| 2 | DLG1 - Q12959 |
| 3 | TP53 - P04637 |
| 4 | MBP - P02686 |
| 5 | RAF1 - P04049 |
| 6 | ARAF - P10398 |
| 7 | TRIM37 - O94972 |
| 8 | SMAD4 - Q13485 |
Regulation
Activation:
Activated by phosphorylation
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
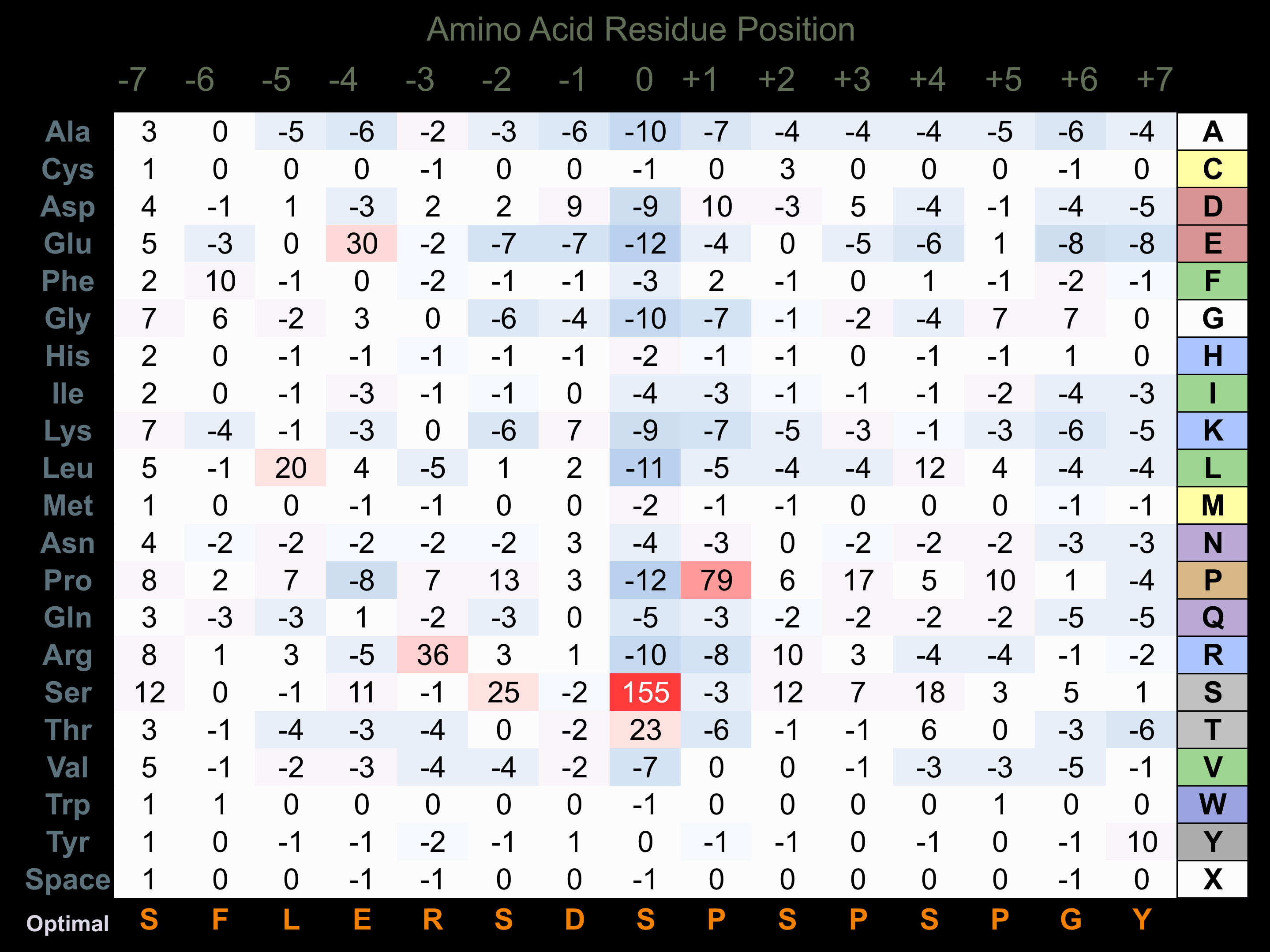
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| K-252a; Nocardiopsis sp. | IC50 < 25 nM | 3813 | 281948 | 22037377 |
| Staurosporine | IC50 > 150 nM | 5279 | 22037377 | |
| BCP9000906 | IC50 > 250 nM | 5494425 | 21156 | 22037377 |
| SB218078 | IC50 > 250 nM | 447446 | 289422 | 22037377 |
| GSK-3b Inhibitor II | IC50 > 1 µM | 6539732 | 288064 | 22037377 |
| KRN633 | IC50 > 1 µM | 406381 | 22037377 | |
| PDK1/Akt/Flt Dual Pathway Inhibitor | IC50 > 1 µM | 5113385 | 599894 | 22037377 |
| PKR Inhibitor | IC50 > 1 µM | 6490494 | 235641 | 22037377 |
| Syk Inhibitor II | IC50 > 1 µM | 16760670 | 22037377 | |
| Y-27632 | IC50 > 1 µM | 448042 | 36228 | 22037377 |
| SureCN4875304 | IC50 > 3.5 µM | 46871765 | 20472445 |
Disease Linkage
General Disease Association:
Cancer, eye disorders
Specific Diseases (Non-cancerous):
Corneal edema
Comments:
Corneal edema is an eye disease characterized by the accumulation of an excessive amount of fluid in the cornea of the eye, potentially leading to damage to the epithelium or endothelium and decreasing visual acuity.
Specific Cancer Types:
Plexiform neurofibromas
Comments:
PBK may be an oncoprotein (OP). Gain-of-function mutations in PBK are linked to cancer, specifically plexiform neuroblastoma, as an overactive PBK protein impairs the G2/M checkpoint and prevents cell cycle arrest in the presence of DNA damage. Despite being absent from most normal somatic tissues, PBK has been shown to be significantly upregulated in several cancer types, indicating that gain-of-function mutations may be causing aberrant PBK expression leading to the augmentation of tumour growth in several forms of human cancer.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +77, p<0.032); Breast non-basal-like cancer (BLC) (%CFC= +129, p<0.0001); Breast sporadic basal-like cancer (BLC) (%CFC= +162, p<0.0001); Cervical cancer (%CFC= -63, p<0.0001); Classical Hodgkin lymphomas (%CFC= +67, p<0.004); Colon mucosal cell adenomas (%CFC= +228, p<0.0001); Large B-cell lymphomas (%CFC= +89, p<0.0005); Ovary adenocarcinomas (%CFC= +502, p<0.061); Prostate cancer - metastatic (%CFC= +233, p<0.002); Prostate cancer - primary (%CFC= +121, p<0.058); Skin melanomas - malignant (%CFC= +210, p<0.0001); and Vulvar intraepithelial neoplasia (%CFC= +299, p<0.0004). The COSMIC website notes an up-regulated expression score for PBK in diverse human cancers of 556, which is 1.2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 36 for this protein kinase in human cancers was 0.6-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.03 % in 24751 diverse cancer specimens. This rate is -55 % lower than the average rate of 0.075 % calculated for human protein kinases in general. Such a low frequency of mutation in human cancers is consistent with this protein kinase playing a role as a tumour requiring protein (TRP).
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.21 % in 603 endometrium cancers tested; 0.21 % in 589 stomach cancers tested; 0.17 % in 1270 large intestine cancers tested; 0.13 % in 238 bone cancers tested; 0.07 % in 1316 breast cancers tested; 0.06 % in 548 urinary tract cancers tested; 0.04 % in 864 skin cancers tested; 0.04 % in 1512 liver cancers tested; 0.02 % in 2009 haematopoietic and lymphoid cancers tested; 0.02 % in 1634 lung cancers tested; 0.02 % in 1459 pancreas cancers tested; 0.02 % in 1276 kidney cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R101H (3).
Comments:
No deletions, insertions or complex mutations are noted on the COSMIC website.

