Nomenclature
Short Name:
AlphaK1
Full Name:
Alpha-protein kinase 1
Alias:
- ALPK1
- EC 2.7.11.-
- LAK
- Lymphocyte alpha-kinase
Classification
Type:
Protein-serine/threonine kinase
Group:
Atypical
Family:
Alpha
SubFamily:
NA
Structure
Mol. Mass (Da):
201,272
# Amino Acids:
1244
# mRNA Isoforms:
2
mRNA Isoforms:
138,861 Da (1244 AA; Q96QP1); 129,951 Da (1166 AA; Q96QP1-2)
4D Structure:
NA
1D Structure:
Subfamily Alignment
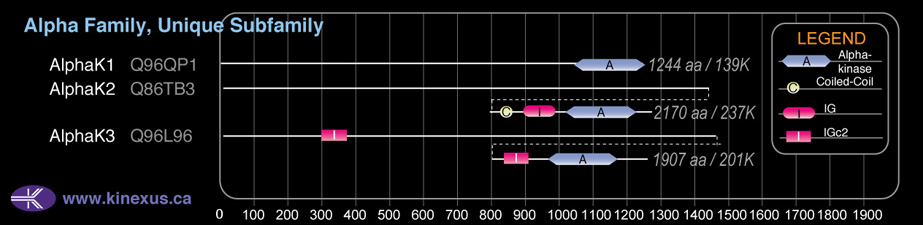
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 291 | 361 | IGc2 |
| 1489 | 1555 | IGc2 |
| 1017 | 1237 | Alpha_kinase |
| 1482 | 1509 | Coiled-coil |
| 1551 | 1634 | IG |
| 1663 | 1883 | Alpha_kinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S124, S716.
Threonine phosphorylated:
T237.
Tyrosine phosphorylated:
Y119, Y717, Y1037.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 87
87
1244
31
1133
 2
2
28
13
31
 3
3
43
5
42
 24
24
341
100
998
 47
47
666
36
535
 3
3
50
55
29
 2
2
28
38
21
 26
26
368
23
579
 29
29
410
13
507
 4
4
64
90
51
 2
2
32
21
31
 51
51
732
95
590
 4
4
56
16
29
 1.5
1.5
21
9
17
 3
3
40
18
49
 2
2
25
17
28
 4
4
64
90
46
 3
3
39
13
49
 3
3
49
80
54
 43
43
613
115
499
 2
2
32
17
39
 3
3
49
18
65
 3
3
49
6
47
 1.3
1.3
19
13
29
 5
5
76
17
121
 64
64
922
64
1419
 3
3
38
22
31
 3
3
39
13
41
 2
2
27
13
35
 3
3
43
42
28
 2
2
32
6
9
 35
35
501
31
645
 3
3
47
115
85
 67
67
960
78
782
 100
100
1431
48
2250
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 98.4
98.4
99.1
95 94.2
94.2
96.1
- -
-
-
76 -
-
-
- 79.9
79.9
87.1
78 -
-
-
- 71.8
71.8
80.9
65 71.8
71.8
80.1
66 -
-
-
- 60.9
60.9
72
- -
-
-
40 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Disease Linkage
General Disease Association:
Inflammatory, and nephrological disorders
Specific Diseases (Non-cancerous):
Gout; Chronic kidney disease (CKD)
Comments:
Gout is a disease characterized by recurrent events of acute inflammatory arthritis most commonly affecting the metatarsal-phalangeal joint of the big toe (~50% of all cases). Associated symptoms may include tophi, kidney stones, or urate nephropathy. Gout is caused by elevated levels of uric acid in the blood, which crystalizes and deposits in joints, tendons, and surrounding tissues. AlphaK3 is a protein-serine/threonine kinase that phosphorylates residues in close association with alpha-helical domains. Single nucleotide polymorphisms in the AlphaK3 gene have been shown to be associated with an elevated risk of developing gout. Three SNPs, namely, rs11726117M861T [C], rs231247 [G], and rs231253 [G,] were the most associated with increased gout risk. Increasing copy numbers of the alleles were correlated with increased risk of gout, indicating an additive model. The associated mRNA displayed reduced hybridization with miR-519e, a microRNA known to inhibit AlphaK3 mRNA expression, indicating a possible dysregulation of AlphaK3 expression. AlphaK3 has also been implicated as a suceptibility gene for chronic kidney disease (CKD) in the Japanese population, particularly in those with diabetes mellitus.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +98, p<0.04); and Skin melanomas - malignant (%CFC= -52, p<0.001). The COSMIC website notes an up-regulated expression score for AlphaK3 in diverse human cancers of 314, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 118 for this protein kinase in human cancers was 2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24726 diverse cancer specimens. This rate is only -3 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.34 % in 805 skin cancers tested; 0.31 % in 1093 large intestine cancers tested; 0.27 % in 602 endometrium cancers tested; 0.18 % in 589 stomach cancers tested; 0.16 % in 1619 lung cancers tested; 0.08 % in 1270 liver cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,010 cancer specimens
Comments:
Only 6 deletions, 1 insertion and no complex mutations are noted on the COSMIC website.

