Nomenclature
Short Name:
NIK
Full Name:
Mitogen-activated protein kinase kinase kinase 14
Alias:
- EC 2.7.11.25
- FTDCR1B
- HS
- HSNIK
- M3K14
- MAP3K14
Classification
Type:
Protein-serine/threonine kinase
Group:
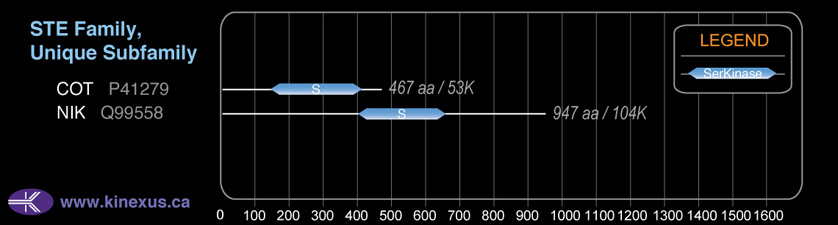
STE
Family:
STE-Unique
SubFamily:
NA
Structure
Mol. Mass (Da):
104042
# Amino Acids:
947
# mRNA Isoforms:
1
mRNA Isoforms:
104,042 Da (947 AA; Q99558)
4D Structure:
Binds to TRAF2, TRAF5, TRAF6, IKKA and NF-kappa-B 2/P100 By similarity. Interacts with PELI3. Interacts with NIBP; the interaction is direct. Interacts with ARRB1 and ARRB2. Interacts with GRB10.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 400 | 687 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S352, S643.
Threonine phosphorylated:
T125, T354, T559+.
Tyrosine phosphorylated:
Y40, Y682.
Ubiquitinated:
K64, K106, K128, K163, K294, K882.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 65
65
706
16
888
 4
4
42
8
40
 11
11
117
15
91
 37
37
401
58
1012
 58
58
627
14
599
 59
59
640
45
1476
 26
26
285
19
481
 43
43
461
31
875
 28
28
303
10
276
 8
8
89
56
77
 7
7
75
26
66
 47
47
511
117
643
 16
16
176
26
194
 3
3
30
6
19
 6
6
69
23
87
 3
3
36
7
13
 16
16
174
112
1575
 9
9
102
20
86
 9
9
93
57
59
 40
40
429
56
461
 6
6
70
22
62
 10
10
110
26
90
 7
7
74
24
89
 8
8
82
20
68
 11
11
114
22
112
 100
100
1080
40
2132
 12
12
134
29
126
 8
8
84
21
77
 7
7
76
20
65
 33
33
352
14
81
 88
88
955
18
622
 50
50
545
21
638
 30
30
322
579
619
 59
59
641
31
652
 22
22
240
22
193
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
99.5 98.1
98.1
98.6
98 -
-
-
89 -
-
-
- 88.6
88.6
92.7
89 -
-
-
- 84.4
84.4
90
85 85.2
85.2
90.3
86 -
-
-
- 32.3
32.3
36.4
- 58.7
58.7
70
63 -
-
-
48 -
-
-
49 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | BIRC2 - Q13490 |
| 2 | MAP2K4 - P45985 |
| 3 | MLXIPL - Q9NP71 |
| 4 | MAPK3 - P27361 |
| 5 | NFKBIA - P25963 |
| 6 | PEBP1 - P30086 |
| 7 | IL1R1 - P14778 |
| 8 | PELI3 - Q8N2H9 |
| 9 | GRB7 - Q14451 |
| 10 | EGFR - P00533 |
| 11 | GRB14 - Q14449 |
| 12 | CASP10 - Q92851 |
| 13 | CASP3 - P42574 |
| 14 | CFLAR - O15519 |
| 15 | RPS15A - P62244 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Ezrin | P15311 | T567 | QGRDKYKTLRQIRQG | |
| IkBa | P25963 | S32 | LLDDRHDSGLDSMKD | - |
| IkBa | P25963 | S36 | RHDSGLDSMKDEEYE | - |
| IKKa (CHUK) | O15111 | S176 | AKDVDQGSLCTSFVG | + |
| IKKa (CHUK) | O15111 | S180 | DQGSLCTSFVGTLQY | + |
| IKKb (IKBKINASE) | O14920 | S177 | AKELDQGSLCTSFVG | + |
| Moesin | P26038 | T558 | LGRDKYKTLRQIRQG | |
| NFkB-p100 | Q00653 | S866 | TAEVKEDSAYGSQSV | + |
| NFkB-p100 | Q00653 | S870 | KEDSAYGSQSVEQEA | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
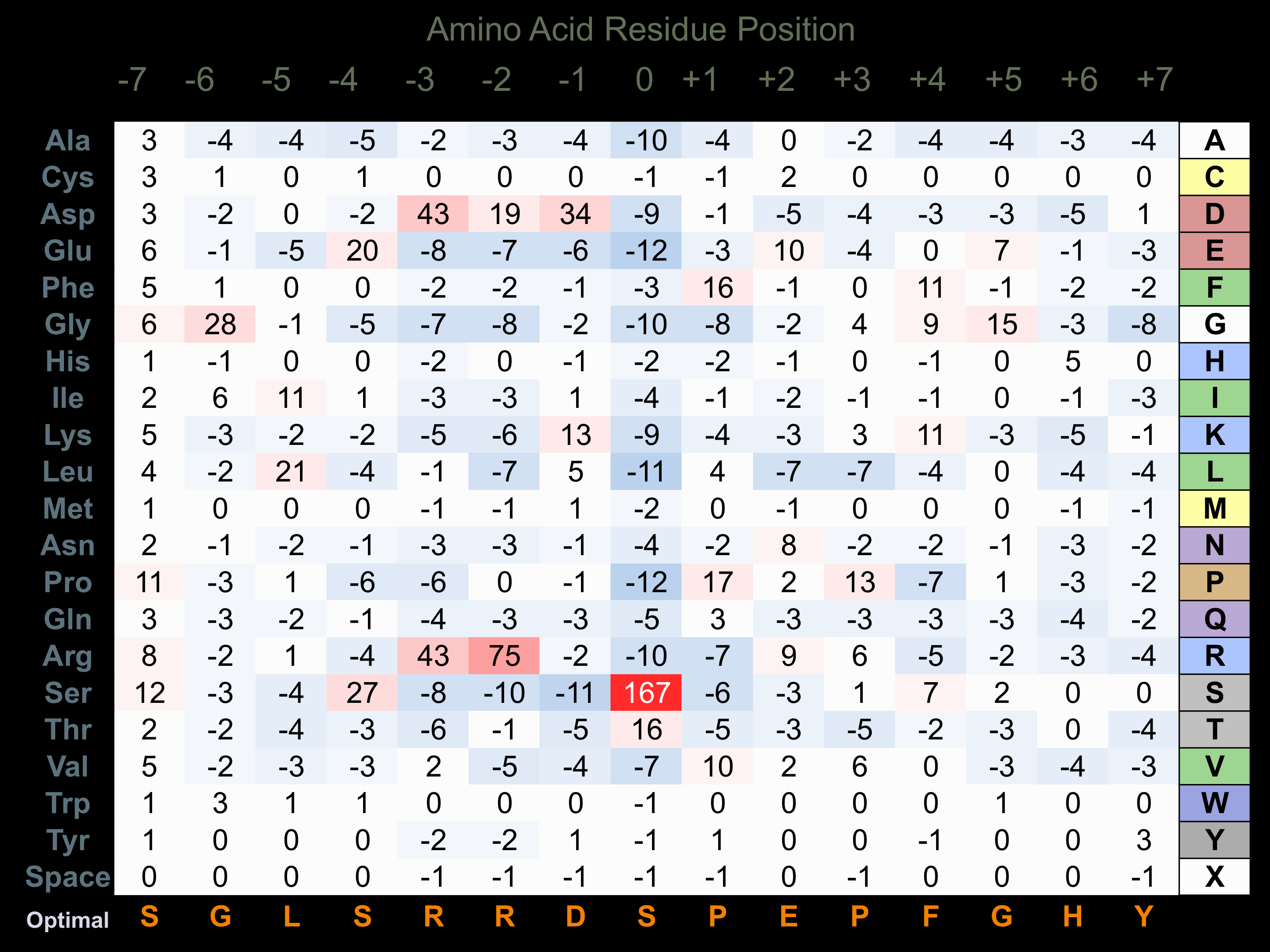
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Cdk1/2 Inhibitor III | IC50 > 150 nM | 5330812 | 261720 | 22037377 |
| K-252a; Nocardiopsis sp. | IC50 = 500 nM | 3813 | 281948 | 22037377 |
| Herbimycin A | IC50 > 1 µM | 6436247 | 1159659 | 22037377 |
| SU4312 | IC50 > 1 µM | 6450842 | 328710 | 22037377 |
| Staurosporine | IC50 = 2.3 µM | 5279 | 20580552 |
Disease Linkage
General Disease Association:
Inflammatory disorders
Specific Diseases (Non-cancerous):
Hypereosinophilic syndrome (HES)
Comments:
Transgenic mice deficient in NIK develop a Hypereosinophilic syndrome (HES)-like disease, reflected by progressive blood and tissue eosinophilia, tissue injury, and premature death at around 25-30 wk of age. Surprisingly, disease development was independent of NIK's activation of I?B kinase a (IKKa) kinase, because mice carrying a mutation in an activatory phosphosite in IKKa, which is targeted by NIK, did not develop inflammatory disease. HES represents a group of inflammatory diseases that is characterized by increased numbers of pathogenic eosinophilic granulocytes in the peripheral blood and diverse organs.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Cervical cancer stage 2B (%CFC= +96, p<0.0009); Colorectal adenocarcinomas (early onset) (%CFC= +86, p<0.001); Ovary adenocarcinomas (%CFC= +71, p<0.042); T-cell prolymphocytic leukemia (%CFC= -51, p<0.01); and Uterine fibroids (%CFC= +88, p<0.007).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 8405 diverse cancer specimens. This rate is only -33 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
None > 3 in 20,705 cancer specimens

