Nomenclature
Short Name:
Wnk3
Full Name:
Serine-threonine-protein kinase WNK3
Alias:
- EC 2.7.11.1
- KIAA1566
- PRKWNK3
- Protein kinase, lysine-deficient 3
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
Wnk
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
191,789
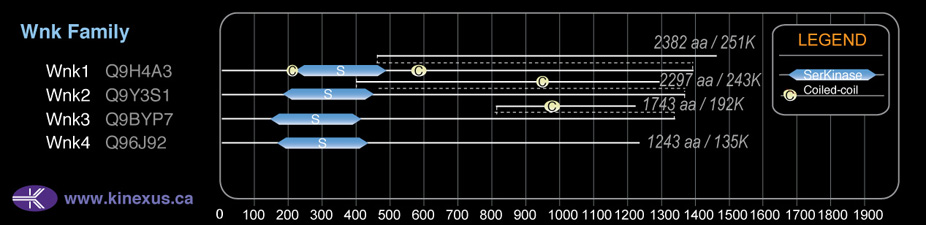
# Amino Acids:
1743
# mRNA Isoforms:
4
mRNA Isoforms:
198,416 Da (1800 AA; Q9BYP7); 197,329 Da (1790 AA; Q9BYP7-2); 192,875 Da (1753 AA; Q9BYP7-4); 191,789 Da (1743 AA; Q9BYP7-3)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 147 | 405 | Pkinase |
| 1484 | 1526 | Coiled-coil |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K439, K443, K468, K1652.
Serine phosphorylated:
S62, S141, S143, S308, S475, S509, S734, S1315, S1471, S1576, S1585, S1638.
Threonine phosphorylated:
T140, T299, T489, T525, T1341.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 54
54
1573
15
1048
 3
3
82
7
93
 -
-
-
-
-
 4
4
102
59
81
 13
13
369
20
228
 0.4
0.4
13
18
8
 0.4
0.4
11
23
7
 83
83
2407
12
4757
 0.1
0.1
2
3
1
 3
3
84
33
84
 1.4
1.4
40
9
66
 18
18
519
21
438
 3
3
95
2
33
 2
2
64
5
102
 1.4
1.4
42
9
66
 2
2
56
12
33
 0.9
0.9
26
141
39
 3
3
97
5
125
 3
3
94
23
150
 13
13
374
56
194
 0.8
0.8
24
9
36
 0.1
0.1
4
7
5
 -
-
-
-
-
 4
4
113
7
108
 0.7
0.7
21
9
35
 25
25
726
41
989
 1.4
1.4
42
5
52
 3
3
76
5
103
 1.3
1.3
38
5
53
 3
3
97
14
82
 20
20
581
24
80
 100
100
2900
18
7599
 12
12
342
50
671
 23
23
678
52
581
 5
5
141
35
91
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.5
99.5
99.8
99 97.8
97.8
98.3
98 -
-
-
86 -
-
-
- 86.7
86.7
91.2
87 -
-
-
- 78.4
78.4
84.9
82 33.6
33.6
48.1
81 -
-
-
- 32.2
32.2
44.8
- 31.2
31.2
44.4
- -
-
-
64 30.3
30.3
40.6
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
38 -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CASP3 - P42574 |
Regulation
Activation:
Activation requires autophosphorylation of Ser-308. Phosphorylation of Ser-304 also promotes increased activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
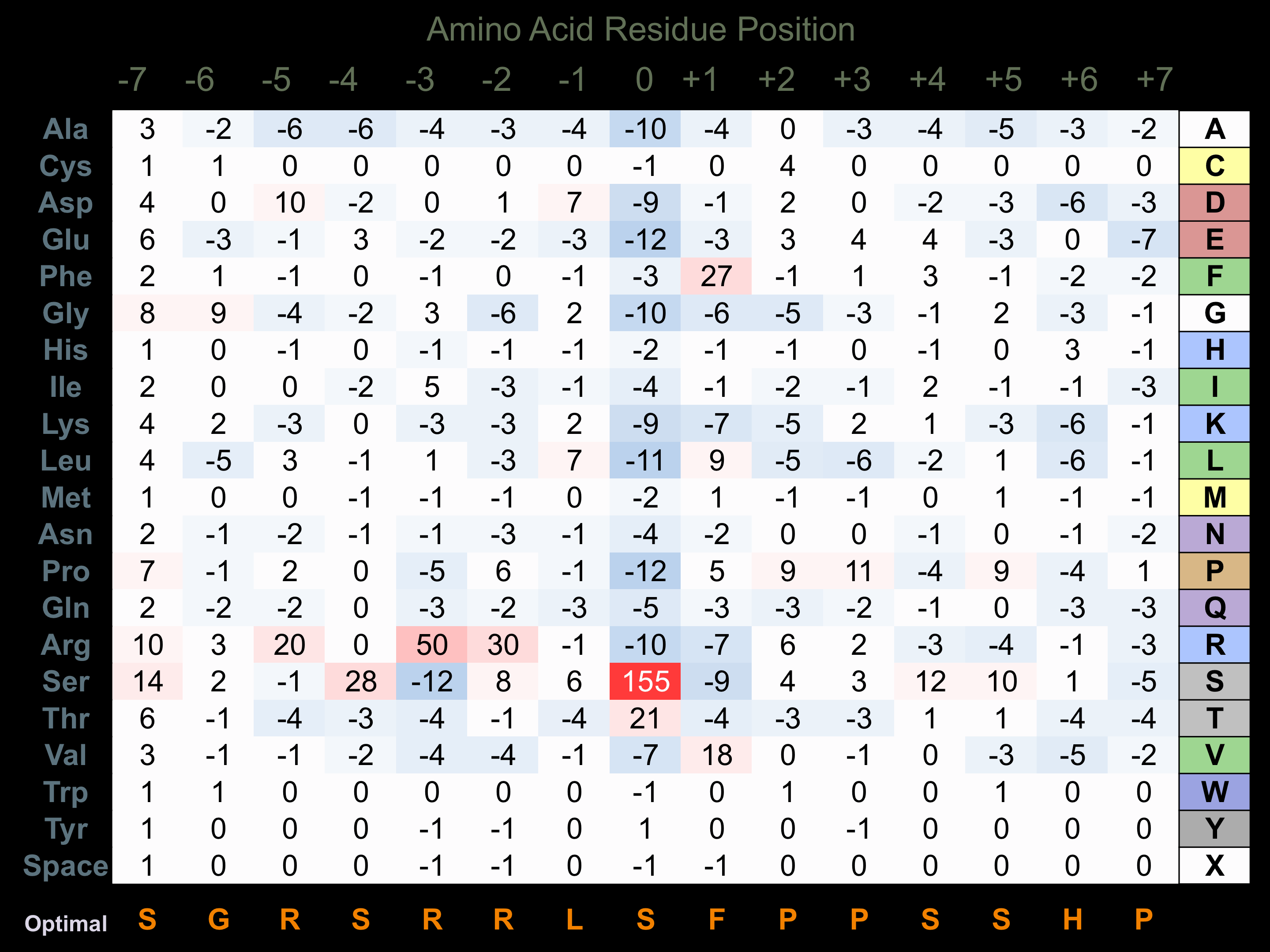
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| AT9283 | IC50 > 100 nM | 24905142 | 19143567 | |
| Vemurafenib | IC50 = 877 nM | 42611257 | 1229517 | 20823850 |
| K-252a; Nocardiopsis sp. | IC50 > 1 µM | 3813 | 281948 | 22037377 |
| MK5108 | IC50 > 1 µM | 24748204 | 20053775 | |
| Silmitasertib | IC50 > 1 µM | 24748573 | 21174434 | |
| SNS314 | IC50 > 1 µM | 16047143 | 514582 | 18678489 |
| JNJ-28871063 | IC50 > 4 µM | 17747413 | 17975007 |
Disease Linkage
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for WNK3 in diverse human cancers of 760, which is 1.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 3 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24898 diverse cancer specimens. This rate is only -7 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.38 % in 1093 large intestine cancers tested; 0.26 % in 805 skin cancers tested; 0.26 % in 602 endometrium cancers tested; 0.19 % in 589 stomach cancers tested; 0.14 % in 1620 lung cancers tested; 0.09 % in 605 oesophagus cancers tested; 0.08 % in 1292 breast cancers tested; 0.05 % in 1270 liver cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: S1393L (6).
Comments:
Only 3 deletions, 3 insertions, and no complex mutations are noted on the COSMIC website.

