Nomenclature
Short Name:
PLK3
Full Name:
Serine-threonine-protein kinase PLK3
Alias:
- CNK
- Kinase PLK3
- PLK-3
- Polo-like kinase 3
- PRK
- Proliferation-related kinase
- Cytokine-inducible serine/threonine-protein kinase
- EC 2.7.11.21
- FGF-inducible kinase
- FNK
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
PLK
SubFamily:
NA
Structure
Mol. Mass (Da):
71,629
# Amino Acids:
646
# mRNA Isoforms:
1
mRNA Isoforms:
71,629 Da (646 AA; Q9H4B4)
4D Structure:
Binds to the calcium/integrin-binding protein (CIB). This interaction probably occurs via the POLO-box domain.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
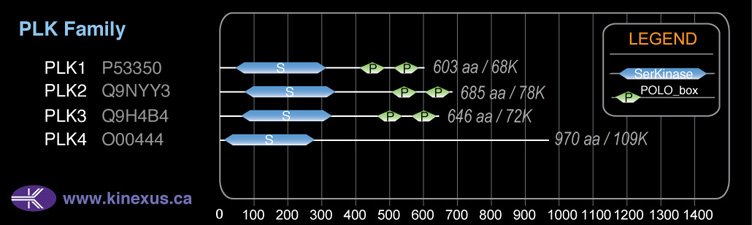
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S510, S515.
Threonine phosphorylated:
T194+, T219+, T223-, T348, T503, T614.
Tyrosine phosphorylated:
Y136+, Y226-, Y506, Y615.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1193
35
1372
 6
6
70
16
66
 7
7
88
16
75
 28
28
331
122
775
 56
56
673
35
622
 41
41
486
98
1516
 15
15
184
41
525
 70
70
832
47
1552
 29
29
347
17
327
 9
9
112
97
176
 6
6
75
35
62
 64
64
758
183
610
 7
7
86
38
92
 6
6
76
12
83
 9
9
110
29
86
 4
4
44
20
36
 23
23
278
128
1213
 7
7
83
25
86
 5
5
65
112
51
 41
41
493
137
512
 9
9
109
27
101
 9
9
102
31
121
 12
12
146
26
131
 11
11
136
25
401
 9
9
109
27
122
 83
83
992
79
1903
 5
5
57
41
45
 11
11
136
25
164
 7
7
88
25
72
 17
17
201
42
96
 53
53
627
24
585
 50
50
596
35
527
 6
6
73
85
126
 70
70
833
83
701
 10
10
122
48
73
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 67.5
67.5
68
0 98.5
98.5
98.6
99 -
-
-
92 -
-
-
93 92.3
92.3
94.9
93 -
-
-
- 91
91
93.5
93.5 89.8
89.8
92.4
94 -
-
-
- 44.3
44.3
57.8
- 65.8
65.8
80.7
68 40.3
40.3
58.5
67 58.7
58.7
76.9
61 -
-
-
- 36.8
36.8
55.9
- 36.7
36.7
56.5
- 35.8
35.8
55.1
- 40.1
40.1
59
- -
-
-
- 28
28
45.2
- -
-
-
- 24.8
24.8
39.9
- 30.8
30.8
48.2
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | TP53 - P04637 |
| 2 | CDC25C - P30307 |
| 3 | MAP2K1 - Q02750 |
| 4 | MAPK1 - P28482 |
| 5 | VRK1 - Q99986 |
| 6 | FZR1 - Q9UM11 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
Cytokine and cellular adhesion trigger FNK induction.
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Cdc25A | P30304 | T80 | RMGSSESTDSGFCLD | |
| Cdc25C | P30307 | S191 | EDQAEEISDELMEFS | |
| Cdc25C | P30307 | S198 | SDELMEFSLKDQEAK | |
| Cdc25C | P30307 | S216 | SGLYRSPSMPENLNR | ? |
| Chk2 (CHEK2) | O96017 | S62 | SSSGTLSSLETVSTQ | |
| Chk2 (CHEK2) | O96017 | S73 | VSTQELYSIPEDQEP | + |
| Cyclin B1 | P14635 | S133 | SPSPMETSGCAPAEE | |
| Jun (c-Jun) | P05412 | S63 | KNSDLLTSPDVGLLK | + |
| Jun (c-Jun) | P05412 | S73 | VGLLKLASPELERLI | + |
| NPM1 | P06748 | S4 | ____MEDSMDMDMSP | |
| p53 | P04637 | S20 | PLSQETFSDLWKLLP | + |
| SNCA | P37840 | S129 | NEAYEMPSEEGYQDY | |
| TOP2A | P11388 | T1343 | FSDFDEKTDDEDFVP | |
| VRK1 | Q99986 | S432 | DDGKLDLSVVENGGL | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
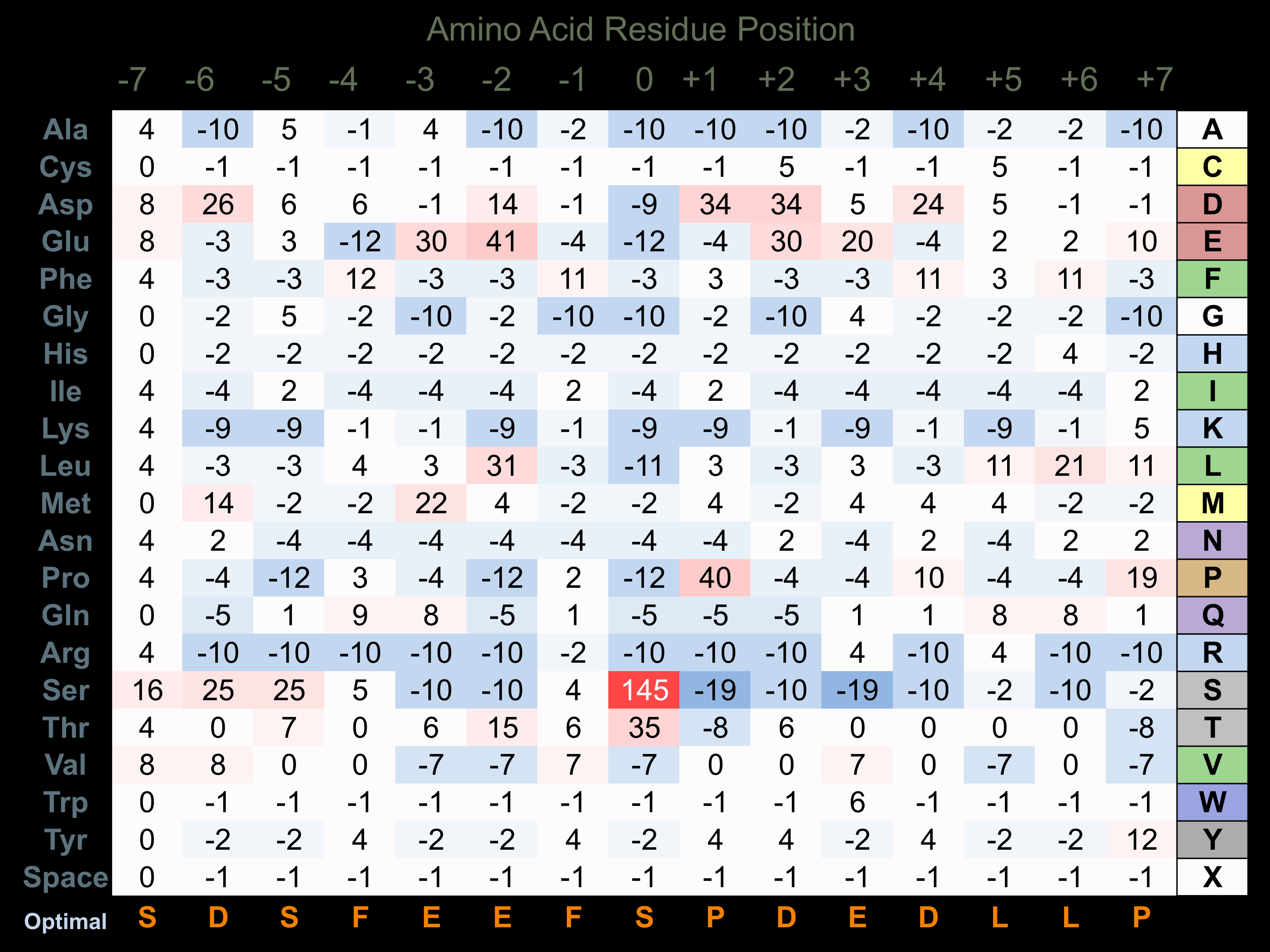
Matrix Type:
Experimentally derived from alignment of 20 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Lung cancer
Comments:
Overexpression of Plk3 leads to mitotic arrest, cytokinesis defects, and apoptosis. Polymorphisms of Plk3 gene were identified in human lung carcinoma cell lines.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for PLK3 in diverse human cancers of 328, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 24743 diverse cancer specimens. This rate is only -20 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.28 % in 1093 large intestine cancers tested; 0.25 % in 805 skin cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,009 cancer specimens
Comments:
Only 9 deletions, and no insertions or complex mutations are noted on the COSMIC website.

