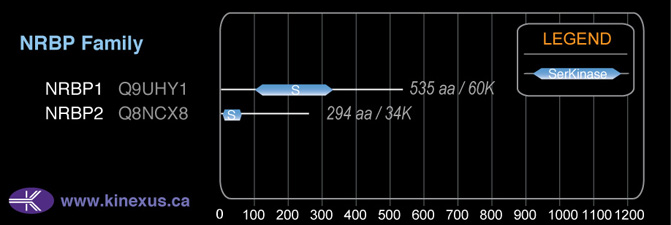
Nomenclature
Short Name:
NRBP2
Full Name:
Nuclear receptor-binding protein 2
Alias:
- Transformation-related gene 16 protein
- TRG-16
- PP9320
- DKFZp434I2411
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
NRBP
SubFamily:
NA
Structure
Mol. Mass (Da):
57,803
# Amino Acids:
501
# mRNA Isoforms:
4
mRNA Isoforms:
57,803 Da (501 AA; Q9NSY0); 36,072 Da (324 AA; Q9NSY0-4); 33,141 Da (293 AA; Q9NSY0-2); 13,910 Da (123 AA; Q9NSY0-5)
4D Structure:
NA
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 38 | 306 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S22, S361, S417.
Threonine phosphorylated:
T409, T411, T419.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 64
64
1184
9
968
 2
2
30
4
30
 -
-
-
-
-
 39
39
713
32
804
 60
60
1104
13
830
 32
32
591
17
567
 3
3
48
13
19
 11
11
212
9
416
 0.7
0.7
12
3
2
 20
20
367
29
258
 48
48
889
5
1164
 35
35
643
22
747
 -
-
-
-
-
 0.9
0.9
17
3
1
 18
18
341
5
433
 6
6
102
5
11
 43
43
789
9
1475
 1
1
22
3
2
 11
11
195
21
99
 59
59
1093
31
570
 90
90
1655
5
2229
 13
13
243
5
323
 -
-
-
-
-
 0.4
0.4
8
3
4
 18
18
336
5
452
 47
47
862
23
687
 0.4
0.4
7
3
1
 0.8
0.8
14
3
3
 1
1
20
3
2
 10
10
182
14
121
 90
90
1667
12
43
 100
100
1847
11
2501
 21
21
380
43
613
 55
55
1007
26
842
 11
11
194
22
172
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 49.4
49.4
50.3
100 43.7
43.7
54.1
- -
-
-
93 -
-
-
- 39.9
39.9
41.4
92 -
-
-
- 94.1
94.1
97.2
94 49.6
49.6
51.3
95 -
-
-
- -
-
-
- 43.4
43.4
49.5
68 31.3
31.3
39
- 31.2
31.2
40.3
66 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
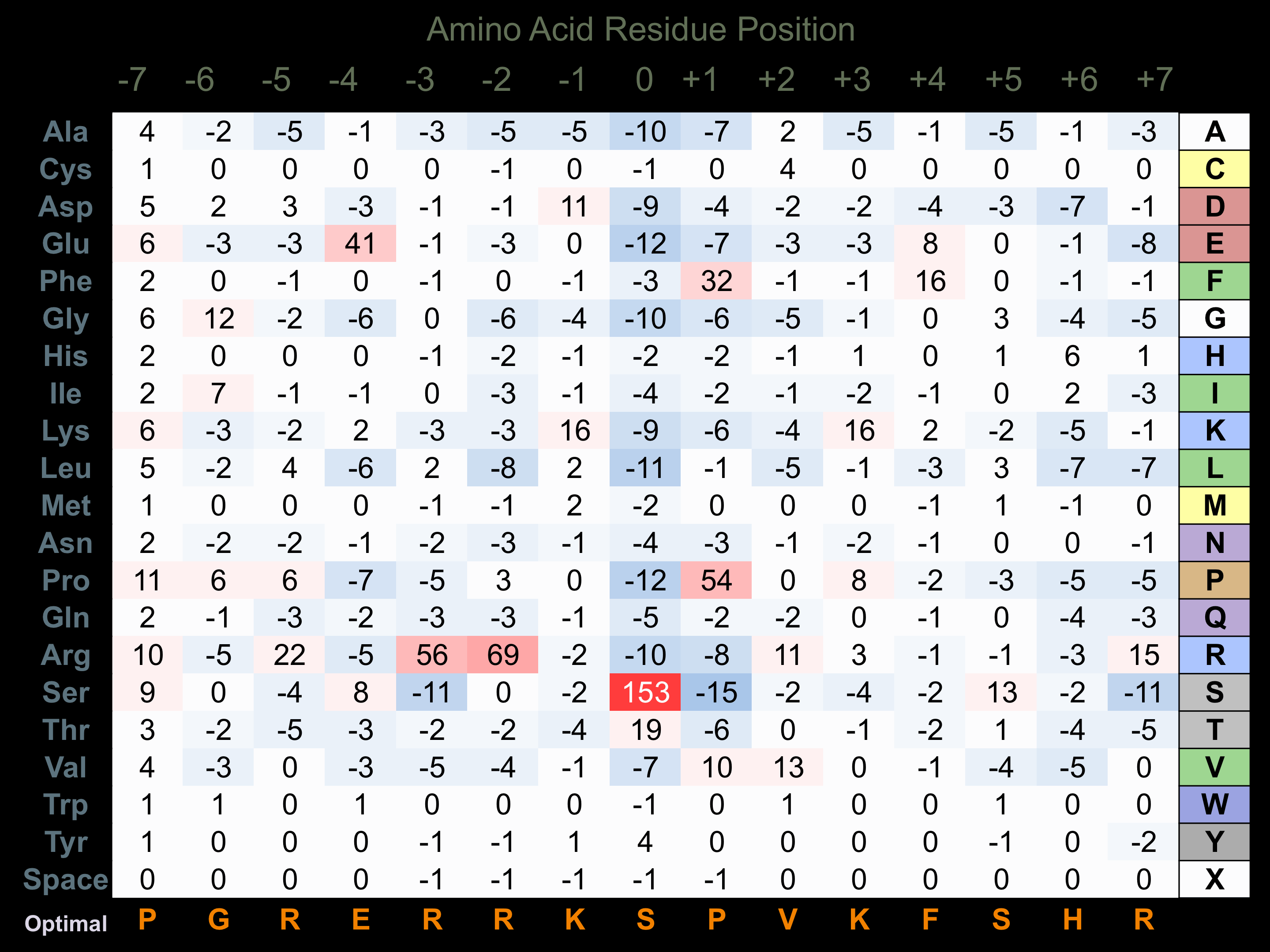
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Medulloblastomas
Comments:
NRBP2 levels are up-regulated by 2-fold in human cancers compared to most protein kinases. NRBP2 is linked to Medulloblastomas, which are a highly malignant form of primary brain cancer that is thought to originate from the aberrant proliferation of neural precursor cells in the granular layers of the cerebellum. It is the most common form of brain tumour in children, accounting for ~16% of all pediatric brain tumours and ~40% of all childhood cerebellar tumours. Medulloblastoma occurence is highest between the ages of 3-4 years and 8-9 years old, following by a decline with increasing age, dropping significantly in adults to account for less than 1% of all reported central nervous system (CNS) tumours. In addition, there is a strong correlation between the occurence of medulloblastoma and Turcot syndrome, as the cancer is observed in ~40% of all Turcot syndrome patients. Elevated NRBP2 expression was reported in a subset of pediatric medulloblastomas, indicating a role for the protein in tumorigenesis. NRBP2 expression is increased following the differentiation of neural stem/progenitor cells in rodents, and has been hypothesized to be involved in the regulation of apoptosis of neural progenitor cells, indicating a potential mechanistic role for its involvement in cancer progression. Analysis of medulloblastoma tissue specimens revealed clusters of NRBP2+ cancer cells that co-expressed neurofillament, but lacked GFAP expression, indicating an association between NRBP2 expression and neuronal differentiation in malignant brain tissue. In addition, the siRNA-mediated knockdown of NRBP2 expression in neural progenitor cells significantly enhanced the vulnerability of the cells to apoptosis. Therefore, elevated expression of NRBP2 is suggested to play an important role in the development of medulloblastoma, potentially by promoting the proliferation of neural progenitor cells and reducing their vulnerability to apoptosis.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for NRBP2 in diverse human cancers of 924, which is 2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 2 for this protein kinase in human cancers was 97% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 24747 diverse cancer specimens. This rate is -40 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.27 % in 589 stomach cancers tested; 0.17 % in 1270 large intestine cancers tested; 0.15 % in 548 urinary tract cancers tested; 0.15 % in 273 cervix cancers tested; 0.13 % in 603 endometrium cancers tested; 0.09 % in 864 skin cancers tested; 0.08 % in 942 upper aerodigestive tract cancers tested; 0.05 % in 1512 liver cancers tested; 0.04 % in 558 thyroid cancers tested; 0.04 % in 1634 lung cancers tested; 0.04 % in 1459 pancreas cancers tested; 0.03 % in 710 oesophagus cancers tested; 0.03 % in 1316 breast cancers tested; 0.02 % in 2103 central nervous system cancers tested; 0.02 % in 2009 haematopoietic and lymphoid cancers tested; 0.02 % in 1276 kidney cancers tested.
Frequency of Mutated Sites:
None > 2 in 20,030 cancer specimens
Comments:
Only 1 insertions, and no deletions or complex mutations are noted on the COSMIC website.

