Nomenclature
Short Name:
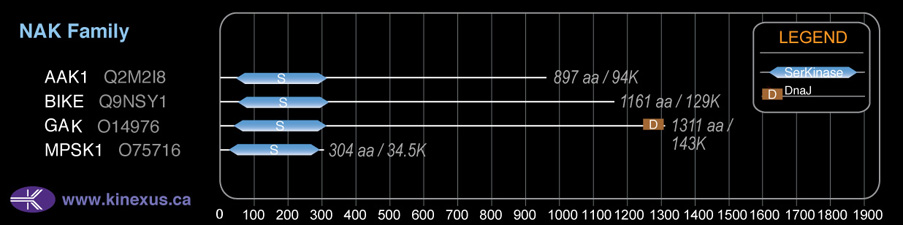
BIKE
Full Name:
BMP-2 inducible protein kinase
Alias:
- BM2K
- BMP2K
- Kinase BIKE
- BMP2 inducible kinase
- DKFZp434K0614
- EC 2.7.11.1
- HRIHFB2017
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
NAK
SubFamily:
NA
Structure
Mol. Mass (Da):
129,172
# Amino Acids:
1161
# mRNA Isoforms:
3
mRNA Isoforms:
129,172 Da (1161 AA; Q9NSY1); 75,812 Da (677 AA; Q9NSY1-3); 73,838 Da (662 AA; Q9NSY1-2)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 51 | 316 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S14, S378, S665, S689, S715, S728, S740, S742, S791, S817, S818, S824, S826, S843, S947, S949, S974, S979, S980, S1029, S1031, S1032, S1039, S1041, S1076, S1080, S1089, S1107, S1111, S1114.
Threonine phosphorylated:
T383, T394, T829, T834, T935, T1002, T1011, T1014, T1037, T1097.
Tyrosine phosphorylated:
Y257, Y594, Y813, Y820, Y1012.
Ubiquitinated:
K991.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 93
93
1001
70
1204
 2
2
19
27
16
 17
17
185
18
253
 23
23
249
229
535
 70
70
761
65
619
 2
2
21
169
30
 11
11
123
75
328
 100
100
1082
77
2925
 43
43
460
31
340
 4
4
39
183
55
 7
7
72
55
117
 52
52
565
312
499
 16
16
168
62
386
 2
2
17
18
9
 6
6
67
43
101
 2
2
22
39
19
 17
17
189
374
1688
 8
8
87
31
152
 2
2
27
195
49
 50
50
544
271
550
 24
24
262
39
469
 22
22
235
46
463
 13
13
145
22
200
 43
43
466
31
885
 11
11
121
39
202
 57
57
613
143
817
 14
14
148
65
351
 8
8
91
31
147
 11
11
116
31
189
 11
11
124
84
143
 37
37
400
48
396
 47
47
508
76
543
 12
12
130
193
514
 79
79
852
156
705
 8
8
84
96
98
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 80.9
80.9
80.9
100 -
-
-
- -
-
-
87 -
-
-
68 88.5
88.5
91.2
90 -
-
-
- 83.2
83.2
88.2
83 38.5
38.5
50
82 -
-
-
- -
-
-
- 72.5
72.5
80.8
88 -
-
-
68 37.9
37.9
45.6
72 -
-
-
- 26.9
26.9
41.7
- 28.9
28.9
38.4
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 21.4
21.4
38.2
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
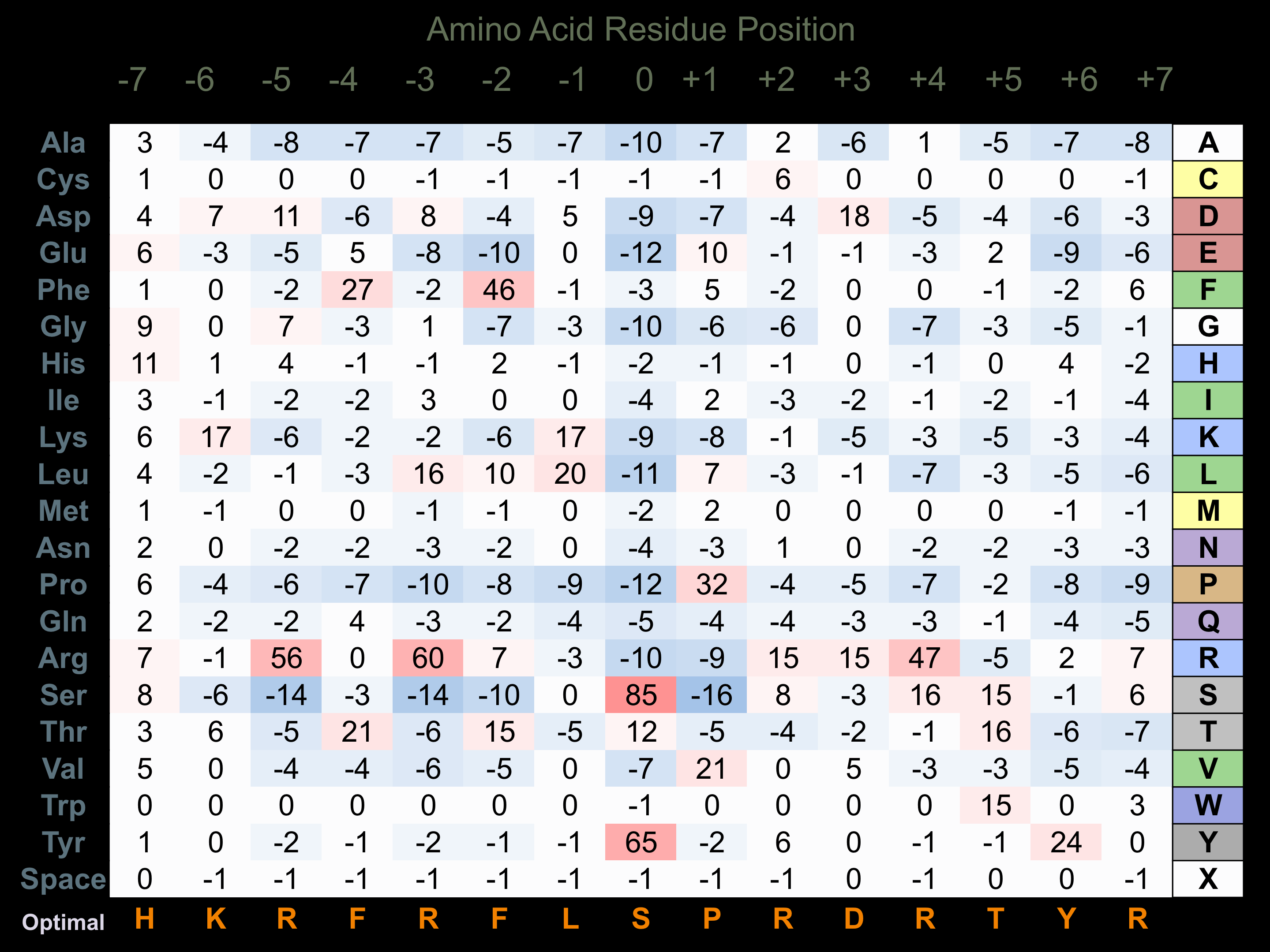
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
Comments:
BIKE may be a tumour requiring protein (TRP), since it displays extremely low rates of mutation in human cancers.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= -85, p<0.043); Brain oligodendrogliomas (%CFC= -72, p<0.088); Breast epithelial cell carcinomas (%CFC= +56, p<0.021); Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= +135, p<0.006); Clear cell renal cell carcinomas (cRCC) (%CFC= +77, p<0.101); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +446, p<0.029); and Ovary adenocarcinomas (%CFC= +49, p<0.03). The COSMIC website notes an up-regulated expression score for BIKE in diverse human cancers of 341, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 5 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.02 % in 24832 diverse cancer specimens. This rate is -80 % lower than the average rate of 0.075 % calculated for human protein kinases in general. Such a very low frequency of mutation in human cancers is consistent with this protein kinase playing a role as a tumour requiring protein (TRP).
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.03 % in 603 endometrium cancers tested; 0.02 % in 864 skin cancers tested; 0.02 % in 1512 liver cancers tested.
Frequency of Mutated Sites:
None > 6 in 20,059 cancer specimens
Comments:
No deletions, insertions or complex mutations are noted on the COSMIC website.

