Nomenclature
Short Name:
TIF1g
Full Name:
Transcription intermediary factor 1-gamma
Alias:
- EC 6.3.2.-
- FLJ11429
- RFG7
- Rfg7 protein
- TRIM33
- Tripartite motif-containing 33 (PTC7;TIF1G)
- KIAA1113
- PTC7
- Ret-fused gene 7
- RET-fused gene 7 protein
Classification
Type:
Protein-serine/threonine kinase
Group:
Atypical
Family:
TIF1
SubFamily:
NA
Structure
Mol. Mass (Da):
122,521
# Amino Acids:
1127
# mRNA Isoforms:
2
mRNA Isoforms:
122,533 Da (1127 AA; Q9UPN9); 120,541 Da (1110 AA; Q9UPN9-2)
4D Structure:
Homooligomer and heterooligomer with TRIM24 and TRIM28 family members. Interacts with SMAD4 in unstimulated cells. Found in a complex with SMAD2 and SMAD3 upon addition of TGF-beta. Interacts with SMAD2 and SMAD3. Interacts with SMAD4 under basal and induced conditions and, upon TGF-beta signaling, with activated SMAD2. Forms a ternary complex with SMAD4 and SMAD2 upon TGF-beta signaling
1D Structure:
Subfamily Alignment
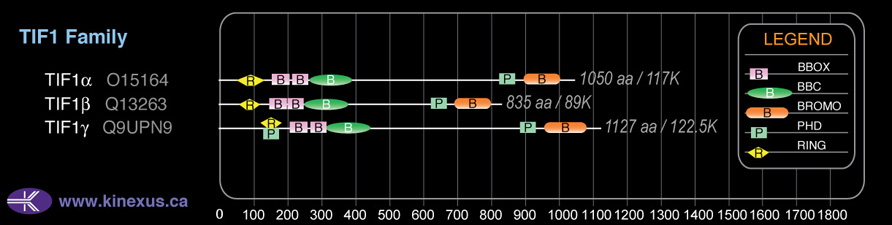
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K252 (N6), K763 (N6), K769 (N6), K776, K793, K861, K950, K951, K953 (N6), K1127, .
Methylated:
R440, R515, R535, R577, R591, R598, R604.
Serine phosphorylated:
S622, S644, S661, S744, S747, S767, S787, S789, S803, S808, S809, S812, S813, S819, S852, S855, S856, S862, S959, S1105, S1119.
Sumoylated:
K776, K793.
Threonine phosphorylated:
T505, T643, T815, T820, T1051, T1102.
Tyrosine phosphorylated:
Y524, Y1048.
Ubiquitinated:
K320, K394.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 41
41
886
58
1131
 3
3
71
27
84
 23
23
486
41
422
 30
30
648
218
914
 43
43
925
47
720
 5
5
113
153
127
 16
16
335
59
566
 100
100
2157
115
4842
 30
30
643
31
514
 8
8
177
186
203
 12
12
251
82
325
 33
33
705
348
669
 11
11
240
85
298
 10
10
208
21
275
 11
11
244
71
264
 4
4
95
29
93
 7
7
153
515
180
 13
13
283
54
287
 6
6
123
188
134
 32
32
685
215
684
 11
11
230
68
230
 17
17
373
74
414
 15
15
328
45
287
 39
39
839
56
952
 14
14
311
68
315
 59
59
1263
143
2067
 12
12
268
88
359
 10
10
207
54
193
 15
15
326
56
323
 3
3
74
56
68
 32
32
686
48
746
 33
33
715
66
679
 15
15
326
146
568
 49
49
1055
104
829
 32
32
700
70
929
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 81.1
81.1
81.1
100 81.3
81.3
81.3
99 -
-
-
98 -
-
-
- 88.9
88.9
90.5
99 -
-
-
- 94.9
94.9
96.3
96 29
29
44.7
96 -
-
-
- 90
90
91.8
- 80.8
80.8
83.4
95 80
80
86.6
89 58.9
58.9
71.1
66 -
-
-
- -
-
-
26 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | NCOR1 - O75376 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Papillary thyroid carcinomas; Follicular thyroid carcinomas
Comments:
TIF1g appears to be an oncoprotein (OP) and a tumour suppressor protein (TSP). Several gain-of-function mutations affecting the TIF1g gene have been observed in cancer patients. For example, a chromosomal translocation involving the TIF1g and RET genes has been linked to the development of thyroid papillary carcinoma. The translocation t(1;10)(p13;q11) generates the TIF1g/RET oncogene, in which RET is fused to the C-terminus of the TIF1g protein, termed the PTC7 rearrangement or oncogene. Another translocation involving RET was also observed in thyroid papillary carcinoma cells, in which RET is fused to the N-terminus of the related TRIM24 gene, termed the PTC6 rearrangement or oncogene. Both chromosomal translocations create oncogenes which promote tumorigenesis in the affected tissues, representing gain-of-function mutations. Interestingly, the occurence of these chromosomal translocations and the associated thyroid papillary carcinoma was observed at a particularly high frequency in susceptible populations after the Chernobyl nuclear reactor accident, indicating a primary role for radiation induced mutagenesis in these translocations and in the pathogenesis of the associated cancer.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +64, p<0.009); Cervical cancer stage 1B (%CFC= -53, p<0.003); Cervical cancer stage 2A (%CFC= -66, p<0.001); Cervical cancer stage 2B (%CFC= -51, p<0.007); and Skin melanomas - malignant (%CFC= -53, p<0.01). The COSMIC website notes an up-regulated expression score for TIF1g in diverse human cancers of 305, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 189 for this protein kinase in human cancers was 3.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 25386 diverse cancer specimens. This rate is only -14 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.39 % in 1093 large intestine cancers tested; 0.28 % in 602 endometrium cancers tested; 0.18 % in 589 stomach cancers tested; 0.17 % in 805 skin cancers tested; 0.12 % in 1270 liver cancers tested; 0.1 % in 1753 lung cancers tested; 0.07 % in 1962 central nervous system cancers tested.
Frequency of Mutated Sites:
None > 5 in 20,655 cancer specimens
Comments:
Only 5 deletions, 4 insertions and no complex mutations are noted on the COSMIC website.

