Nomenclature
Short Name:
Wnk2
Full Name:
Serine-threonine-protein kinase WNK2
Alias:
- Antigen NY-CO-43
- EC 2.7.11.1
- PRKWNK2
- Protein kinase with no lysine 2
- Protein kinase, lysine-deficient 2
- WNK lysine deficient protein kinase 2
- KIAA1760
- NY-CO-43
- P,OKcl.13
- P/OKcl.13
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
Wnk
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
242,676
# Amino Acids:
2297
# mRNA Isoforms:
4
mRNA Isoforms:
242,676 Da (2297 AA; Q9Y3S1); 238,279 Da (2254 AA; Q9Y3S1-4); 234,948 Da (2224 AA; Q9Y3S1-2); 84,868 Da (779 AA; Q9Y3S1-3)
4D Structure:
NA
1D Structure:
Subfamily Alignment
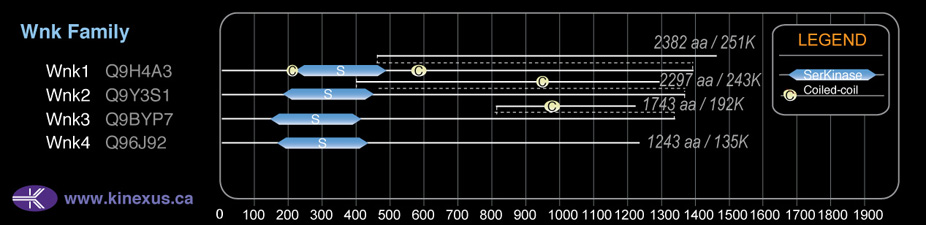
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 195 | 453 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Methylated:
R1631.
Serine phosphorylated:
S45, S49, S352, S356, S560, S1150, S1152, S1262, S1588, S1685, S1689, S1736, S1777, S1798, S1817, S1818, S1862, S1889, S1918, S1936, S2056, S2067, S2110, S2128, S2129.
Threonine phosphorylated:
T13, T232, T347, T1175, T1894, T1908, T2078, T2157, T2159, T2173.
Tyrosine phosphorylated:
Y210, Y442, Y1693, Y1804.
Ubiquitinated:
K349.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1752
36
1658
 19
19
327
12
684
 8
8
136
6
99
 5
5
93
139
144
 42
42
736
43
354
 3
3
53
49
182
 1.4
1.4
24
53
21
 36
36
635
22
986
 0.3
0.3
5
3
1
 3
3
61
65
174
 7
7
125
17
192
 39
39
691
58
601
 6
6
108
10
91
 14
14
245
7
447
 15
15
262
11
251
 2
2
42
29
110
 9
9
157
171
168
 58
58
1012
13
2956
 8
8
139
49
369
 32
32
568
132
466
 11
11
195
17
149
 5
5
86
13
86
 6
6
113
6
88
 14
14
241
17
334
 9
9
154
17
126
 57
57
1007
97
1882
 8
8
135
12
188
 17
17
306
13
406
 5
5
93
13
80
 5
5
81
28
45
 27
27
471
12
29
 80
80
1408
41
3745
 18
18
320
123
696
 44
44
772
130
660
 4
4
77
74
51
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 29.8
29.8
43
97 86.9
86.9
87.9
96 -
-
-
82.5 -
-
-
- 29.2
29.2
42.4
81 -
-
-
- 76.1
76.1
79.9
86 29.8
29.8
43.4
87 -
-
-
- 55
55
64.5
- 54.5
54.5
63.7
68 -
-
-
61 38.8
38.8
50.1
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
40 -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | ABL1 - P00519 |
| 2 | FYN - P06241 |
| 3 | ATXN1 - P54253 |
Regulation
Activation:
Activation requires autophosphorylation of Ser-356. Phosphorylation of Ser-352 also promotes increased activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
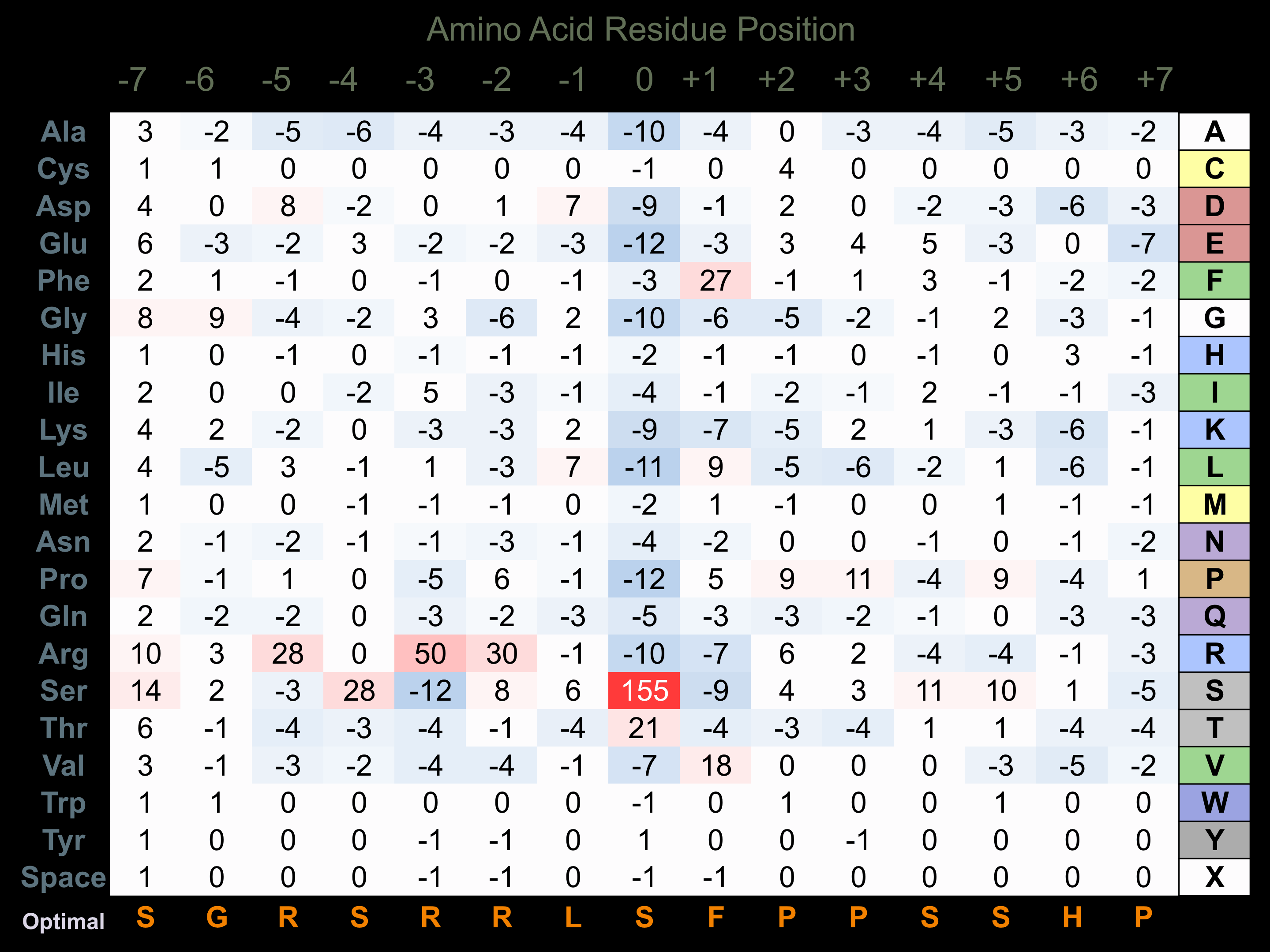
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| AT9283 | IC50 > 100 nM | 24905142 | 19143567 | |
| LY364947 | IC50 = 500 nM | 447966 | 261454 | 22037377 |
| MK5108 | IC50 > 1 µM | 24748204 | 20053775 | |
| Silmitasertib | IC50 > 1 µM | 24748573 | 21174434 | |
| SNS314 | IC50 > 1 µM | 16047143 | 514582 | 18678489 |
| CHEMBL248757 | Ki > 1.518 µM | 44444843 | 248757 | 17935989 |
| JNJ-28871063 | IC50 > 4 µM | 17747413 | 17975007 |
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Colon cancer
Comments:
WNK2 may be a tumour suppressor protein (TSP). Wnk2 is also called serologically defined colon cancer antigen 43. Loss of Wnk2 expression was found in adult gliomas and triggers Rac1-mediated tumour cell invasiveness.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= -47, p<0.012); and Ovary adenocarcinomas (%CFC= +103, p<0.0002). The COSMIC website notes an up-regulated expression score for WNK2 in diverse human cancers of 322, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 25158 diverse cancer specimens. This rate is only -23 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.3 % in 805 skin cancers tested; 0.27 % in 1093 large intestine cancers tested; 0.18 % in 589 stomach cancers tested; 0.12 % in 602 endometrium cancers tested; 0.11 % in 814 upper aerodigestive tract cancers tested; 0.08 % in 1753 lung cancers tested; 0.07 % in 1270 liver cancers tested; 0.04 % in 1226 kidney cancers tested; 0.03 % in 1962 central nervous system cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: V1786G (22).
Comments:
At least 20 deletionsmutations have been identified spread over the full length of the protein.

