Nomenclature
Short Name:
KHS1
Full Name:
Mitogen-activated protein kinase kinase kinase kinase 5
Alias:
- EC 2.7.11.1
- GCKR
- M4K5
- MAP4K5
- MAPKKKK5
- Germinal center kinase-related
- Germinal center kinase-related enzyme
- KHS
- Kinase KHS1
Classification
Type:
Protein-serine/threonine kinase
Group:
STE
Family:
STE20
SubFamily:
KHS
Structure
Mol. Mass (Da):
95,040
# Amino Acids:
846
# mRNA Isoforms:
1
mRNA Isoforms:
95,040 Da (846 AA; Q9Y4K4)
4D Structure:
Interacts with both SH3 domains of the adapter proteins CRK and CRKL.
1D Structure:
Subfamily Alignment
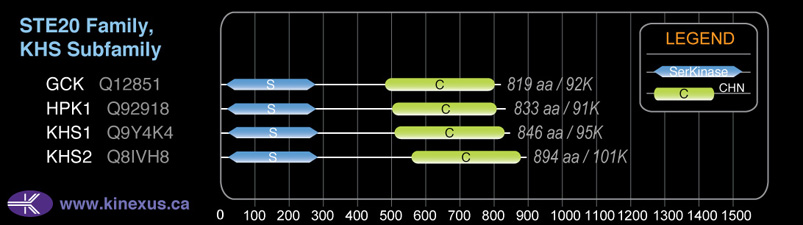
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S174+, S335, S399, S400, S433, S434, S436, S445+.
Threonine phosphorylated:
T30, T168+, T178-, T267, T304, T320.
Tyrosine phosphorylated:
Y20, Y31-, Y35, Y303, Y401, Y558.
Ubiquitinated:
K500.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 39
39
1020
42
1143
 0.8
0.8
20
20
14
 18
18
473
33
402
 15
15
389
158
430
 31
31
818
35
687
 27
27
709
119
2601
 6
6
159
43
332
 50
50
1320
80
2587
 24
24
640
24
507
 4
4
118
129
206
 4
4
117
56
137
 23
23
603
240
559
 5
5
142
66
194
 2
2
52
15
37
 6
6
166
49
192
 2
2
47
22
27
 9
9
228
154
1413
 12
12
306
44
317
 4
4
108
148
177
 27
27
703
162
749
 22
22
574
46
634
 12
12
309
51
382
 13
13
335
44
362
 8
8
199
44
225
 9
9
226
46
248
 27
27
706
108
1157
 10
10
257
69
380
 9
9
248
44
272
 27
27
704
44
773
 4
4
107
42
76
 24
24
634
30
688
 100
100
2627
57
6750
 20
20
530
90
1014
 33
33
880
83
753
 3
3
84
48
68
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.7
99.7
99.8
100 25.3
25.3
39.8
99 -
-
-
97.5 -
-
-
98 98.1
98.1
99
98 -
-
-
- 96
96
98.2
96 68.7
68.7
81.3
97 -
-
-
- 87.1
87.1
90.1
- 24.6
24.6
39.7
91 25.4
25.4
40.3
82 25.1
25.1
40.5
79 -
-
-
- 25.9
25.9
43.3
- -
-
-
- 23.8
23.8
41.1
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CRKL - P46109 |
| 2 | CRK - P46108 |
| 3 | TRAF2 - Q12933 |
| 4 | ABL1 - P00519 |
| 5 | TANK - Q92844 |
| 6 | BCR - P11274 |
| 7 | GRB2 - P62993 |
| 8 | NCK1 - P16333 |
| 9 | FYN - P06241 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
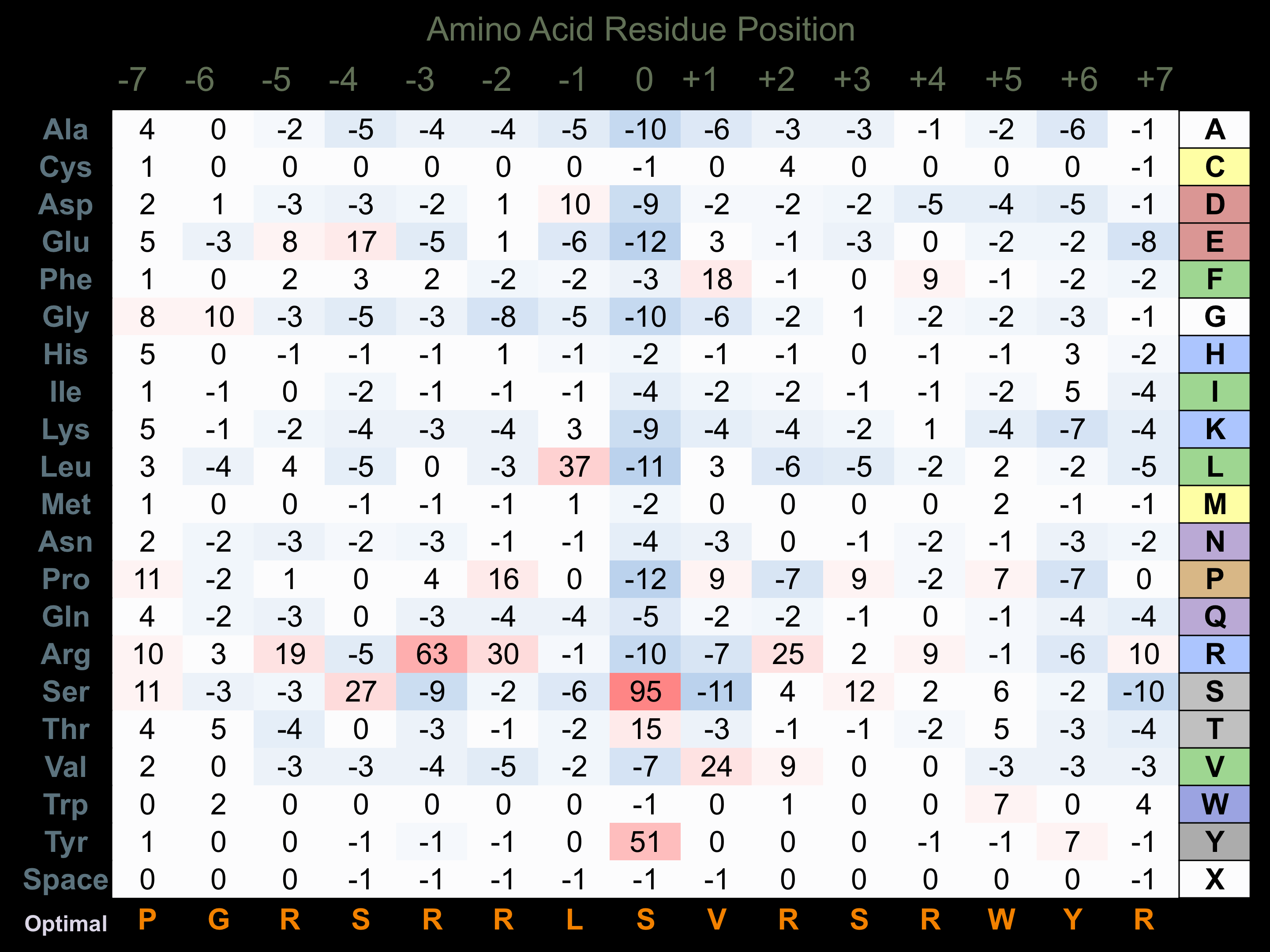
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= -46, p<0.015); Bladder carcinomas (%CFC= +114, p<0.0002); Cervical cancer (%CFC= +63, p<0.001); and Malignant pleural mesotheliomas (MPM) tumours (%CFC= +86, p<0.0008). The COSMIC website notes an up-regulated expression score for KHS1 in diverse human cancers of 415, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 183 for this protein kinase in human cancers was 3.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis. A K49R mutation inhibits its kinase phosphotransferase activity and inhibits downstream JNK activation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.04 % in 24939 diverse cancer specimens. This rate is -42 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.25 % in 1093 large intestine cancers tested; 0.2 % in 602 endometrium cancers tested; 0.18 % in 805 skin cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,222 cancer specimens
Comments:
Only 6 deletions and 2 insertions, and no complex mutations are noted on the COSMIC website.

