Nomenclature
Short Name:
RSK3
Full Name:
Ribosomal protein S6 kinase alpha 1
Alias:
- 90 kDa ribosomal protein S6 kinase 2
- EC 2.7.11.1
- Ribosomal protein S6 kinase alpha 2
- Ribosomal protein S6 kinase, 90kDa, polypeptide 2
- RPS6KA2
- S6K-alpha; S6K-alpha 2
- HU-2
- KS6A2
- MAPKAPK1C
- Pp90RSK3
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
RSK
SubFamily:
RSK
Specific Links
Structure
Mol. Mass (Da):
83239
# Amino Acids:
733
# mRNA Isoforms:
3
mRNA Isoforms:
85,532 Da (758 AA; Q15349-2); 84,014 Da (741 AA; Q15349-3); 83,239 Da (733 AA; Q15349)
4D Structure:
Forms a complex with either ERK1 or ERK2 in quiescent cells. Transiently dissociates following mitogenic stimulation
1D Structure:
Subfamily Alignment
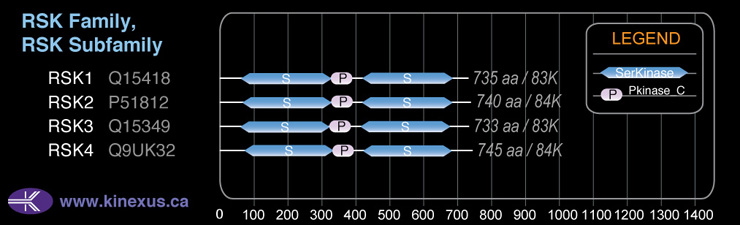
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K91, K94, K295.
Other:
K630 (Glycyl lysine isopeptide (Lys-Gly) in interchain with G-Cter in ubiquitin).
Serine phosphorylated:
S26, S27, S28, S30, S106, S206, S218-, S377, S381, S382, S388, S546, S628, S650.
Threonine phosphorylated:
T222-, T471, T570, T574-, T618, T662.
Tyrosine phosphorylated:
Y135, Y217+, Y225-, Y426, Y463, Y540, Y573+.
Ubiquitinated:
K630.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1153
38
1264
 9
9
109
18
67
 8
8
87
22
72
 45
45
517
130
540
 85
85
979
32
670
 5
5
59
83
44
 22
22
252
45
466
 40
40
461
55
629
 37
37
426
17
291
 10
10
120
106
103
 5
5
56
43
53
 71
71
815
203
738
 7
7
81
44
30
 8
8
94
15
54
 11
11
122
19
84
 13
13
154
21
101
 17
17
198
285
1349
 8
8
92
32
80
 13
13
150
103
95
 61
61
703
132
612
 24
24
280
39
185
 12
12
138
42
95
 7
7
82
32
53
 4
4
44
32
50
 5
5
54
39
38
 54
54
625
85
704
 5
5
61
47
29
 6
6
72
33
62
 11
11
126
33
63
 23
23
265
28
172
 55
55
639
36
459
 41
41
475
41
529
 9
9
103
113
436
 71
71
822
83
694
 16
16
186
48
192
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
95 84.7
84.7
86.2
- -
-
-
97 -
-
-
98 93.9
93.9
95.2
97 -
-
-
- 95.6
95.6
97.8
96 77.1
77.1
87.6
96 -
-
-
- 80.3
80.3
87.8
- 76.4
76.4
86.4
94 75
75
84.9
87.5 75.8
75.8
85.9
89 -
-
-
- 23.9
23.9
37.7
- -
-
-
- 56.7
56.7
71.5
60 -
-
-
- -
-
-
- -
-
-
- -
-
-
48 -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Activated by multiple phosphorylations on threonine and serine residues. Phosphorylation at Ser-360 increases phosphotransferase activity. Phosphorylation at Ser-218 inhibits interaction with AKAP6.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
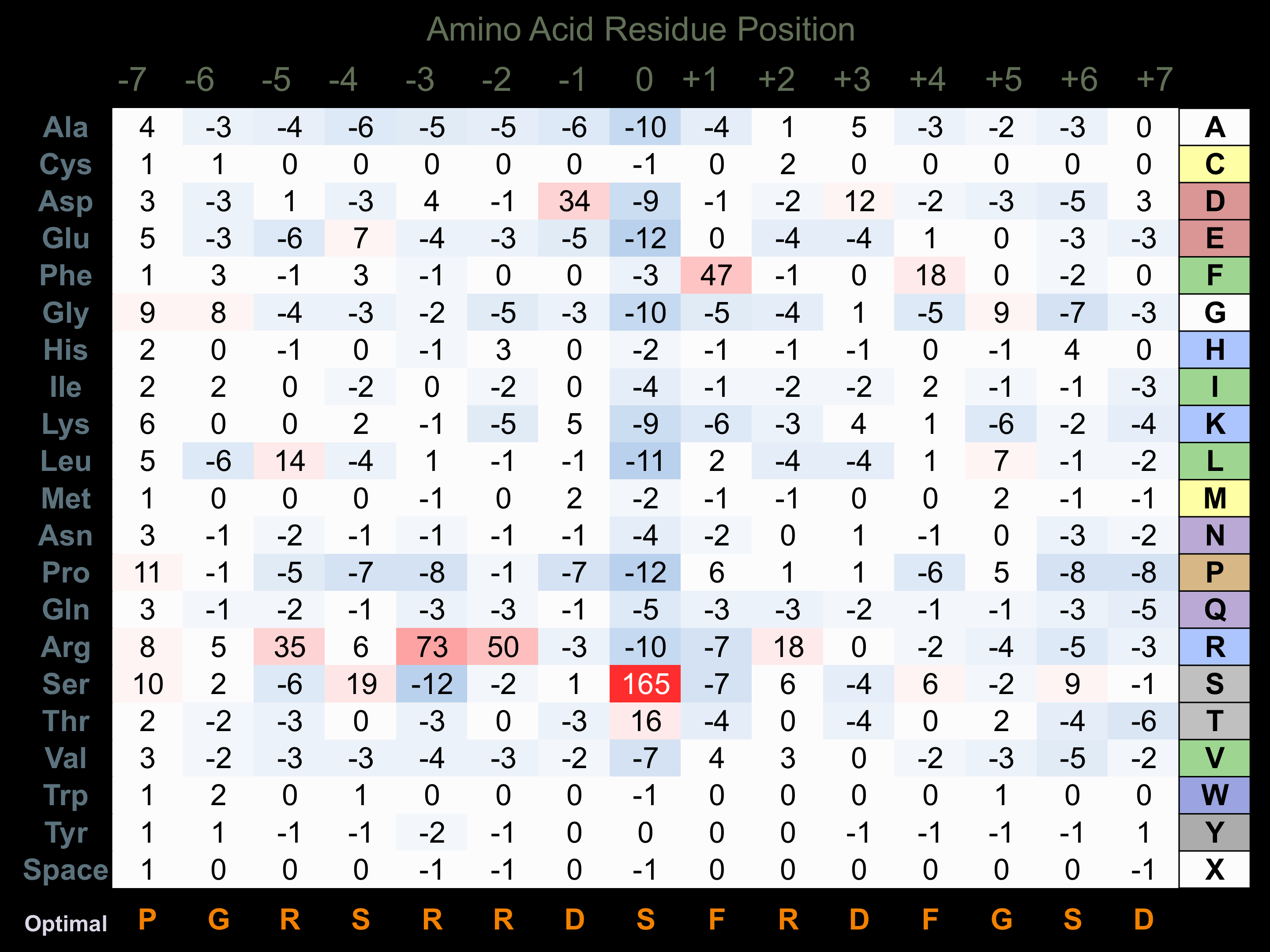
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
2
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cognitive impairment
Specific Diseases (Non-cancerous):
Coffin-Lowry syndrome (CLS)
Comments:
The rare syndrome Coffin-Lowry Syndrome (CLS) is characterized by cognitive impairment, short stature, head, face, and skeletal abnormalities (including upper jaw deformation and curvature of the spine), broad nose, extensive brow, eyelid folds that are slanting downwards, widely-spaced eyes, and thick eyebrows. CLS can lead to issues eating, respiratory issues, cognitive impairment, developmental delay, impaired hearing, strange gait, stimulus-induced drop episodes, and heart or kidney issues.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Colorectal adenocarcinomas (early onset) (%CFC= +93, p<0.001); Prostate cancer - primary (%CFC= +92, p<0.0001); and Vulvar intraepithelial neoplasia (%CFC= -48, p<0.002). The COSMIC website notes an up-regulated expression score for RSK3 in diverse human cancers of 287, which is 0.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 28 for this protein kinase in human cancers was 0.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.1 % in 25630 diverse cancer specimens. This rate is only 33 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.75 % in 1093 large intestine cancers tested; 0.37 % in 589 stomach cancers tested; 0.36 % in 805 skin cancers tested; 0.36 % in 602 endometrium cancers tested; 0.11 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
None > 5 in 20,841 cancer specimens
Comments:
Only 2 deletions, 1 insertion, and no complex mutations are noted on the COSMIC website.

