Nomenclature
Short Name:
SNRK
Full Name:
Alias:
- FLJ20224
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
CAMKL
SubFamily:
SNRK
Structure
Mol. Mass (Da):
84,276
# Amino Acids:
765
# mRNA Isoforms:
2
mRNA Isoforms:
84,276 Da (765 AA; Q9NRH2); 27,481 Da (244 AA; Q9NRH2-2)
4D Structure:
NA
1D Structure:
Subfamily Alignment
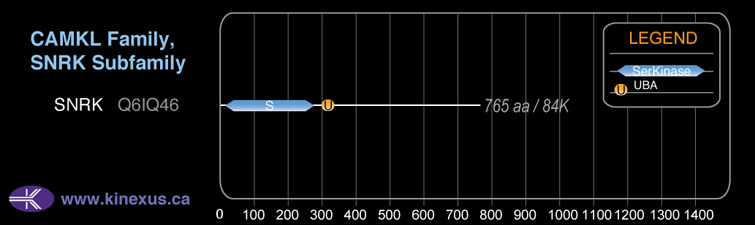
Domain Distribution:
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K11 (N6), K68.
Serine phosphorylated:
S162, S174, S275, S351, S362, S390, S460, S495, S518, S531, S536, S537, S550, S563, S569, S570, S587, S607.
Threonine phosphorylated:
T157, T172, T173+, T380, T463, T521, T543, T617, T619, T623.
Tyrosine phosphorylated:
Y562.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 76
76
924
35
810
 4
4
52
18
64
 20
20
247
3
198
 31
31
371
109
505
 60
60
729
32
655
 11
11
130
90
161
 22
22
265
45
433
 37
37
450
35
597
 63
63
759
17
688
 12
12
143
87
178
 7
7
85
24
110
 52
52
626
192
671
 14
14
167
25
163
 5
5
62
15
86
 7
7
85
19
96
 8
8
94
21
117
 22
22
267
202
2288
 5
5
55
14
67
 11
11
132
84
138
 61
61
731
132
769
 9
9
107
16
128
 18
18
217
22
301
 8
8
91
21
115
 8
8
98
14
132
 13
13
159
18
218
 58
58
696
66
693
 9
9
110
28
125
 8
8
92
13
122
 13
13
162
13
202
 6
6
72
28
59
 100
100
1208
24
810
 56
56
678
35
657
 40
40
480
86
910
 64
64
768
88
732
 30
30
364
48
582
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.6
99.6
99.6
100 99.4
99.4
99.6
99.5 -
-
-
95 -
-
-
- 91.9
91.9
93.4
94 -
-
-
- 89.8
89.8
92.9
92 89.8
89.8
92.4
92 -
-
-
- 93.8
93.8
96.4
- 25.8
25.8
45.2
92 -
-
-
87 22.3
22.3
38.2
78 -
-
-
- 41.6
41.6
56
50 38
38
49.3
- 21.8
21.8
36.7
31 -
-
-
- -
-
-
- -
-
-
- -
-
-
- 26.1
26.1
41
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Activated by phosphorylation on Thr-173 by STK11 in complex with STE20-related adapter-alpha (STRAD alpha) pseudo kinase and CAB39.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| H3.3 | P84243 | S10 | TKQTARKSTGGKAPR | |
| Trehalose-6-phosphate synthase iso 5 (Arabidopsis) | O23617 | S22 | SGNFHSFSREKKRFP | |
| Trehalose-6-phosphate synthase iso 5 (Arabidopsis) | O23617 | S49 | DDDNNSNSVCSDAPS |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
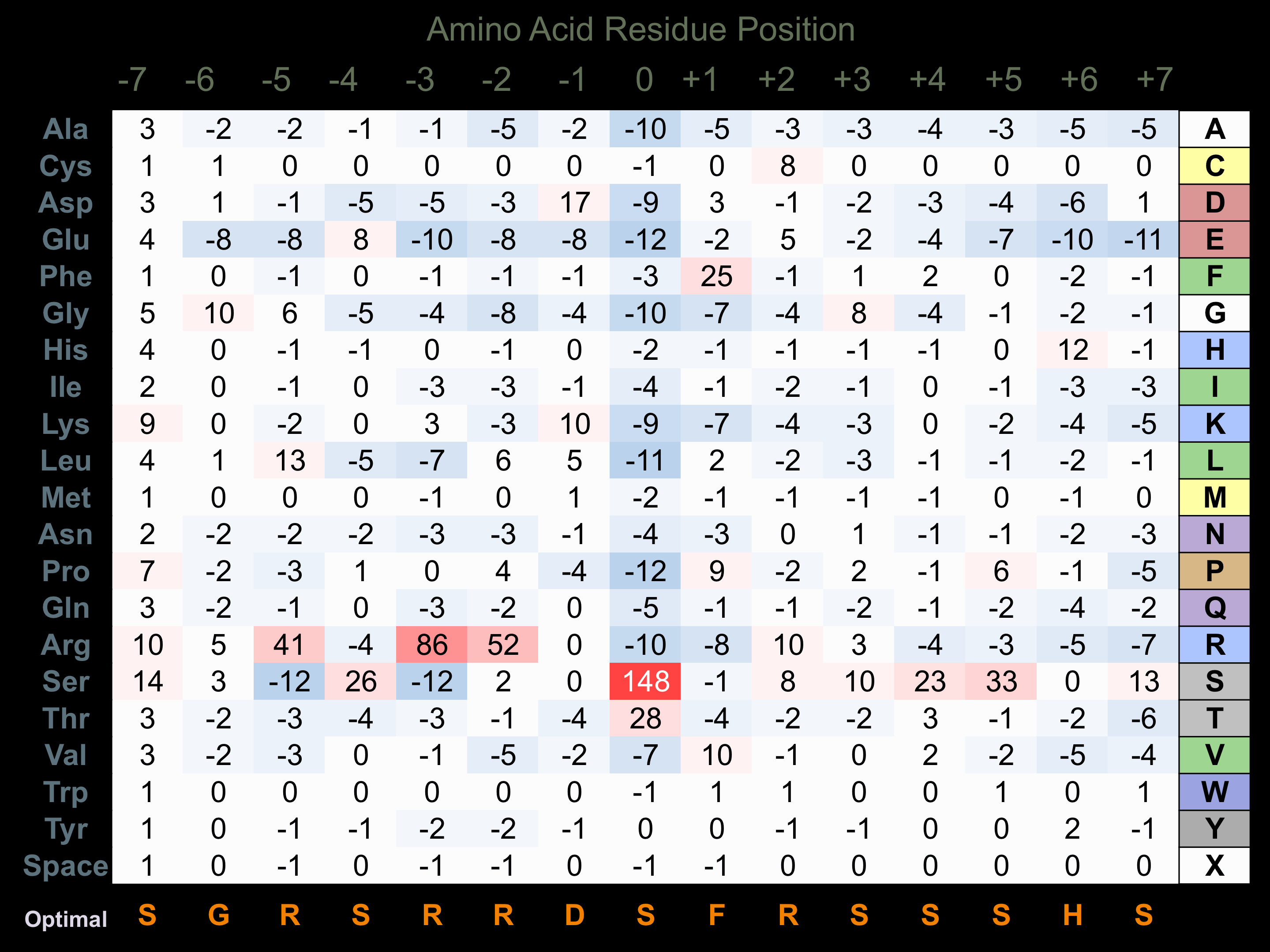
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Staurosporine | Kd = 90 nM | 5279 | 22037378 | |
| AT9283 | IC50 > 100 nM | 24905142 | 19143567 | |
| TG101348 | Kd = 230 nM | 16722836 | 1287853 | 22037378 |
| NVP-TAE684 | Kd = 300 nM | 16038120 | 509032 | 22037378 |
| Sunitinib | Kd = 640 nM | 5329102 | 535 | 22037378 |
| Silmitasertib | IC50 > 1 µM | 24748573 | 21174434 | |
| SNS314 | IC50 > 1 µM | 16047143 | 514582 | 18678489 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| SU14813 | Kd = 1.5 µM | 10138259 | 1721885 | 22037378 |
| JNJ-28871063 | IC50 > 4 µM | 17747413 | 17975007 |
Disease Linkage
Comments:
SNRK is a protein-serine/threonine kinase that may play a role in hematopoietic cell proliferation or differentiation, and is also a potential potential mediator of neuronal apoptosis. T173A substitution is associated with less phosphorylation and activation. R248Q and S147P mutations in SNRK may link with susceptibility to the development of vascular anomalies in humans.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +105, p<0.064); Cervical cancer (%CFC= +118, p<0.026); Lung adenocarcinomas (%CFC= -54, p<0.0001); Oral squamous cell carcinomas (OSCC) (%CFC= -62, p<0.056); and Skin melanomas - malignant (%CFC= -57, p<0.02). The COSMIC website notes an up-regulated expression score for SNRK in diverse human cancers of 255, which is 0.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 102 for this protein kinase in human cancers was 1.7-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. A T173A substitution can result in reduced phosphorylation and activation of SNRK. R248Q and S147P mutations may link with susceptibility to the development of vascular anomalies in humans.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24782 diverse cancer specimens. This rate is only -6 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.32 % in 1270 large intestine cancers tested; 0.29 % in 589 stomach cancers tested; 0.27 % in 864 skin cancers tested; 0.2 % in 603 endometrium cancers tested; 0.16 % in 1512 liver cancers tested; 0.11 % in 833 ovary cancers tested; 0.11 % in 238 bone cancers tested; 0.07 % in 1316 breast cancers tested; 0.06 % in 1459 pancreas cancers tested; 0.05 % in 548 urinary tract cancers tested; 0.05 % in 273 cervix cancers tested; 0.04 % in 710 oesophagus cancers tested; 0.04 % in 1634 lung cancers tested; 0.03 % in 942 upper aerodigestive tract cancers tested; 0.03 % in 441 autonomic ganglia cancers tested; 0.02 % in 558 thyroid cancers tested; 0.02 % in 1276 kidney cancers tested; 0.01 % in 881 prostate cancers tested; 0.01 % in 2082 central nervous system cancers tested; 0.01 % in 2009 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R310* (4).
Comments:
Only 2 deletions, and no insertions or complex mutations are noted on the COSMIC website.

