Nomenclature
Short Name:
SgK110
Full Name:
Similar to protein kinase Bsk146
Alias:
- SgK110 (first kinase domain) with SgK069 (2nd kinase domain)
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
NKF1
SubFamily:
NA
Structure
Mol. Mass (Da):
38,488
# Amino Acids:
359
# mRNA Isoforms:
1
mRNA Isoforms:
38,488 Da (359 AA; P0C264)
4D Structure:
NA
1D Structure:
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 43 | 306 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-

 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 66
66
70.6
97 75.3
75.3
76.8
- -
-
-
83.5 -
-
-
- 31.7
31.7
36.3
83 -
-
-
- 83.1
83.1
87.8
87 39.5
39.5
53
85 -
-
-
- 28.5
28.5
35.2
- 31.3
31.3
44
48 -
-
-
38 32.9
32.9
47.5
- -
-
-
- -
-
-
- 29.1
29.1
42.9
- -
-
-
- 31.8
31.8
46.7
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
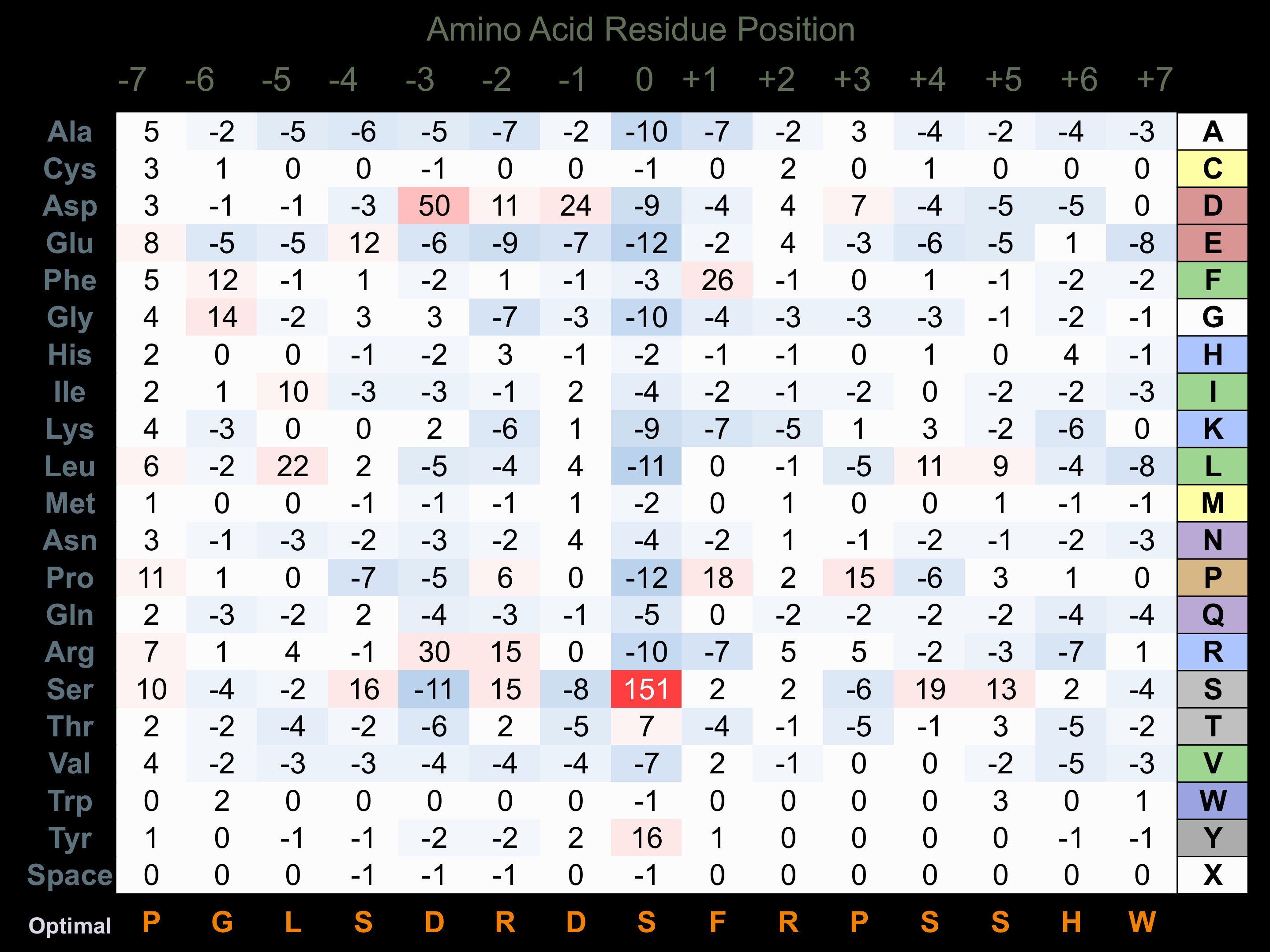
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Staurosporine | Kd = 25 nM | 5279 | 22037378 | |
| NVP-TAE684 | Kd = 54 nM | 16038120 | 509032 | 22037378 |
| Lestaurtinib | Kd = 120 nM | 126565 | 22037378 | |
| R406 | Kd = 190 nM | 11984591 | 22037378 | |
| Aurora A Inhibitor 1 (DF) | Kd < 200 nM | 21992004 | ||
| KW2449 | Kd = 230 nM | 11427553 | 1908397 | 22037378 |
| Crizotinib | Kd = 240 nM | 11626560 | 601719 | 22037378 |
| Aurora A Inhibitor 23 (DF) | Kd < 400 nM | 21992004 | ||
| Tozasertib | Kd = 530 nM | 5494449 | 572878 | 22037378 |
| Aurora A Inhibitor 29 (DF) | Kd < 800 nM | 21992004 | ||
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| JNJ-28312141 | Kd = 1.1 µM | 22037378 | ||
| PD173955 | Kd = 1.1 µM | 447077 | 386051 | 22037378 |
| Sunitinib | Kd = 1.9 µM | 5329102 | 535 | 19654408 |
| Nintedanib | Kd = 2 µM | 9809715 | 502835 | 22037378 |
| TG101348 | Kd = 2.9 µM | 16722836 | 1287853 | 22037378 |
| BMS-690514 | Kd < 4.5 µM | 11349170 | 21531814 |
Disease Linkage
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Frequency of Mutated Sites:
None > 1 in 19,905 cancer specimens

