Nomenclature
Short Name:
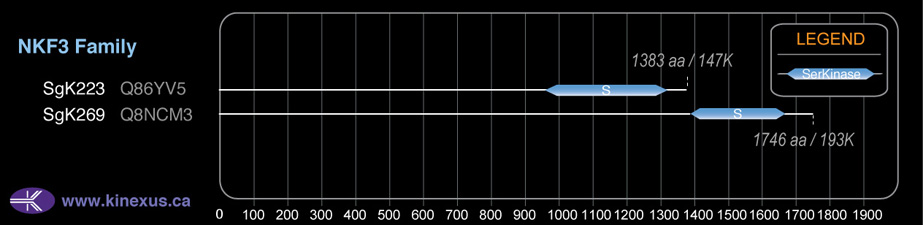
SgK269
Full Name:
KIAA2002 protein (partial sequence)
Alias:
- EC 2.7.10.2
- FLJ21140
- FLJ34483
- KIAA2002
- SG269
- Tyrosine-protein kinase SgK269: Sugen kinase 269
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
NKF3
SubFamily:
NA
Structure
Mol. Mass (Da):
193,106
# Amino Acids:
1746
# mRNA Isoforms:
1
mRNA Isoforms:
193,106 Da (1746 AA; Q9H792)
4D Structure:
NA
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 1329 | 1667 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K563, K567, K581, K921.
Serine phosphorylated:
S281, S359, S370, S377, S389, S495, S503, S507, S527, S540, S568, S572, S587, S643, S648, S725, S779, S786, S792, S812, S819, S826, S837, S847, S854, S856, S861, S864, S877, S878, S898, S927, S1035, S1036, S1038, S1046, S1057, S1104, S1214, S1217, S1250, S1253, S1256, S1347, S1374, S1563, S1657.
Threonine phosphorylated:
T215, T293, T504, T510, T515, T539, T571, T574, T576, T615, T657, T783, T813, T822, T834, T852, T860, T876, T900, T937, T1074, T1077, T1078, T1106, T1151, T1165, T1583.
Tyrosine phosphorylated:
Y387, Y462, Y475, Y531, Y598, Y616, Y635+, Y641, Y665, Y797, Y880, Y1107, Y1215, Y1348, Y1371, Y1372, Y1587.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 66
66
1010
25
1214
 2
2
37
6
49
 0.9
0.9
14
1
0
 29
29
442
67
502
 68
68
1041
20
682
 3
3
48
46
95
 8
8
122
22
132
 52
52
784
19
868
 20
20
303
7
198
 14
14
207
54
216
 4
4
55
9
60
 71
71
1086
66
725
 0.7
0.7
10
12
3
 1.2
1.2
19
3
5
 6
6
93
6
119
 3
3
47
12
44
 16
16
240
75
149
 3
3
43
3
32
 11
11
171
52
224
 44
44
663
81
668
 17
17
257
5
328
 39
39
588
7
982
 0.9
0.9
13
2
4
 2
2
25
3
11
 16
16
236
5
310
 98
98
1495
41
2916
 0.5
0.5
8
12
5
 1.4
1.4
21
3
8
 1.1
1.1
16
3
5
 10
10
150
28
85
 0.6
0.6
9
6
3
 100
100
1522
26
3183
 64
64
970
98
1508
 66
66
1006
52
816
 28
28
431
35
277
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.5
99.5
99.8
100 56
56
56.4
98 -
-
-
91 -
-
-
- 66.1
66.1
69.5
83 -
-
-
- 85.9
85.9
90.8
87 86.1
86.1
90.8
87 -
-
-
- 76.8
76.8
85
- 70.3
70.3
79.8
74 27.2
27.2
42.5
54 52.2
52.2
65.8
57 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
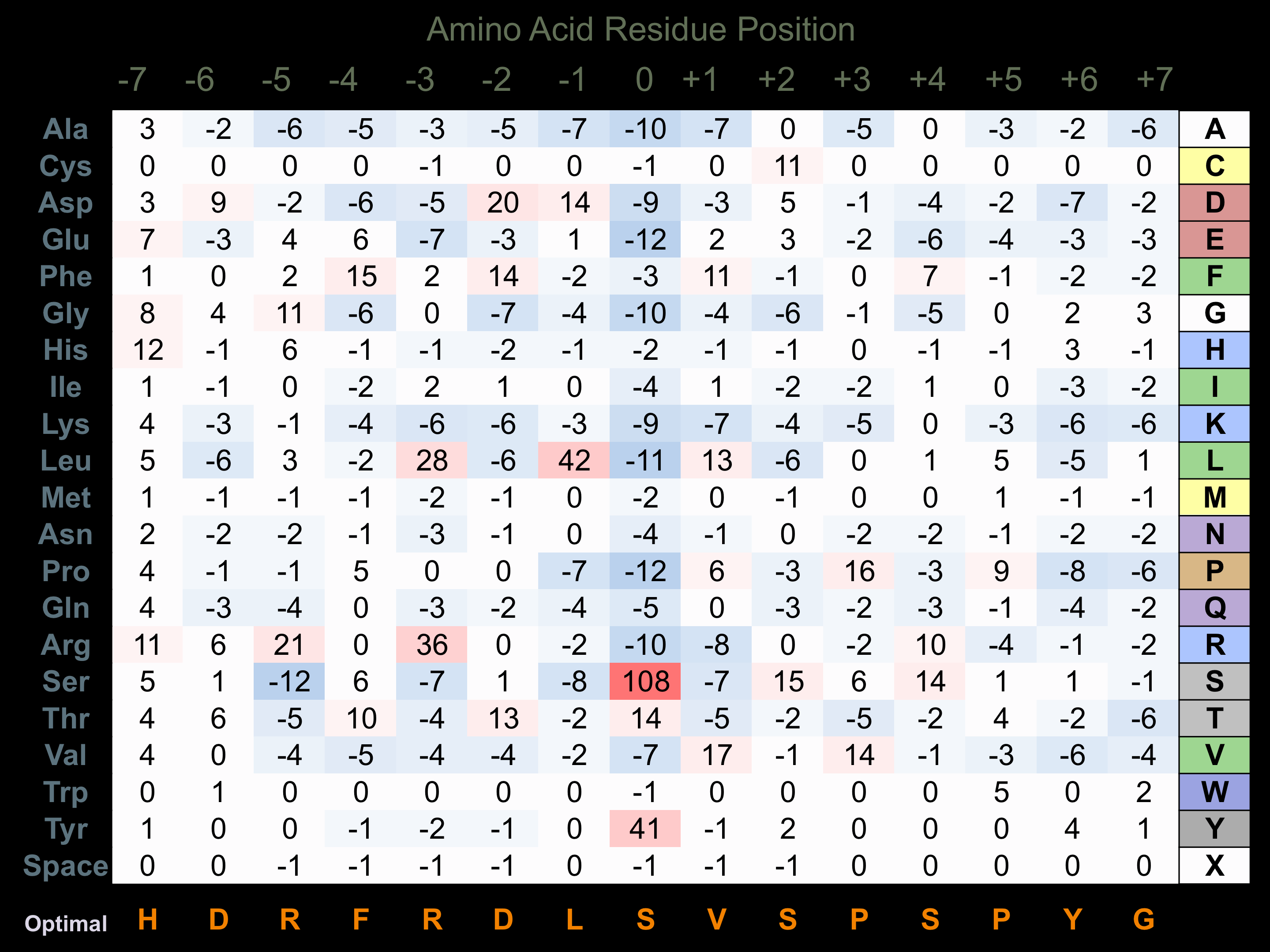
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Pancreatic ductal adenocarcinomas (PDAC)
Comments:
SgK269 has also been demonstrated to function downstream of eIF5A1 and eIF5A2, translation initiation factors that have been implicated in several forms of human cancer. For example, the expression of these factors is required for the growth of pancreatic ductal adenocarcinoma (PDAC) cells in vivo and their overexpression in mice models leads to enhanced PDAC cell growth and tumour formation. Downstream activity of PEAK1 is required for the PDAC growth promoting effects of the eIF5A factors, indicating a role for SgK269 in the tumorigenesis of PDAC.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= -55, p<0.0005); and Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +4391, p<0.0004).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 24702 diverse cancer specimens. This rate is only -20 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.15 % in 569 stomach cancers tested; 0.14 % in 1229 large intestine cancers tested; 0.1 % in 864 skin cancers tested; 0.1 % in 603 endometrium cancers tested; 0.09 % in 65 Meninges cancers tested; 0.08 % in 273 cervix cancers tested; 0.05 % in 807 ovary cancers tested; 0.05 % in 548 urinary tract cancers tested; 0.04 % in 942 upper aerodigestive tract cancers tested; 0.04 % in 710 oesophagus cancers tested; 0.04 % in 1608 lung cancers tested; 0.03 % in 1512 liver cancers tested; 0.01 % in 881 prostate cancers tested; 0.01 % in 558 thyroid cancers tested; 0.01 % in 441 autonomic ganglia cancers tested; 0.01 % in 382 soft tissue cancers tested; 0.01 % in 2169 haematopoietic and lymphoid cancers tested; 0.01 % in 2074 central nervous system cancers tested; 0.01 % in 1289 breast cancers tested; 0 % in 1253 kidney cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R297Q (3); R297* (3); H611N (2); H611V (2); H611R; H611Q (1).
Comments:
Only 3 deletions, and no insertions or complex mutations are noted on the COSMIC website.

