Nomenclature
Short Name:
SgK494
Full Name:
Sugen kinase 494
Alias:
- AK057735
- EC=2.7.11.1
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
RSK
SubFamily:
NA
Structure
Mol. Mass (Da):
31044
# Amino Acids:
274
# mRNA Isoforms:
1
mRNA Isoforms:
46,223 Da (410 AA; L7N484)
4D Structure:
NA
1D Structure:
Subfamily Alignment
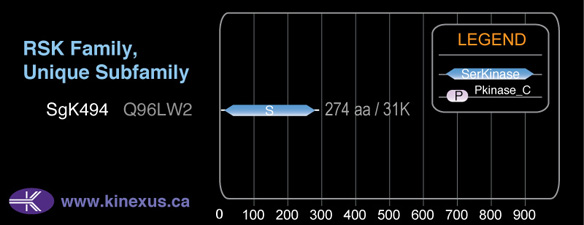
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 107 | 359 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-

 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 98.9
98.9
99.2
99 93.4
93.4
96.3
- -
-
-
87.5 -
-
-
- 58.5
58.5
62
86 -
-
-
- 82
82
88.4
86 -
-
-
85 -
-
-
- -
-
-
- -
-
-
- -
-
-
62 -
-
-
56 -
-
-
- -
-
-
- -
-
-
- 22.2
22.2
34.9
42 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PSTPIP1 - O43586 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
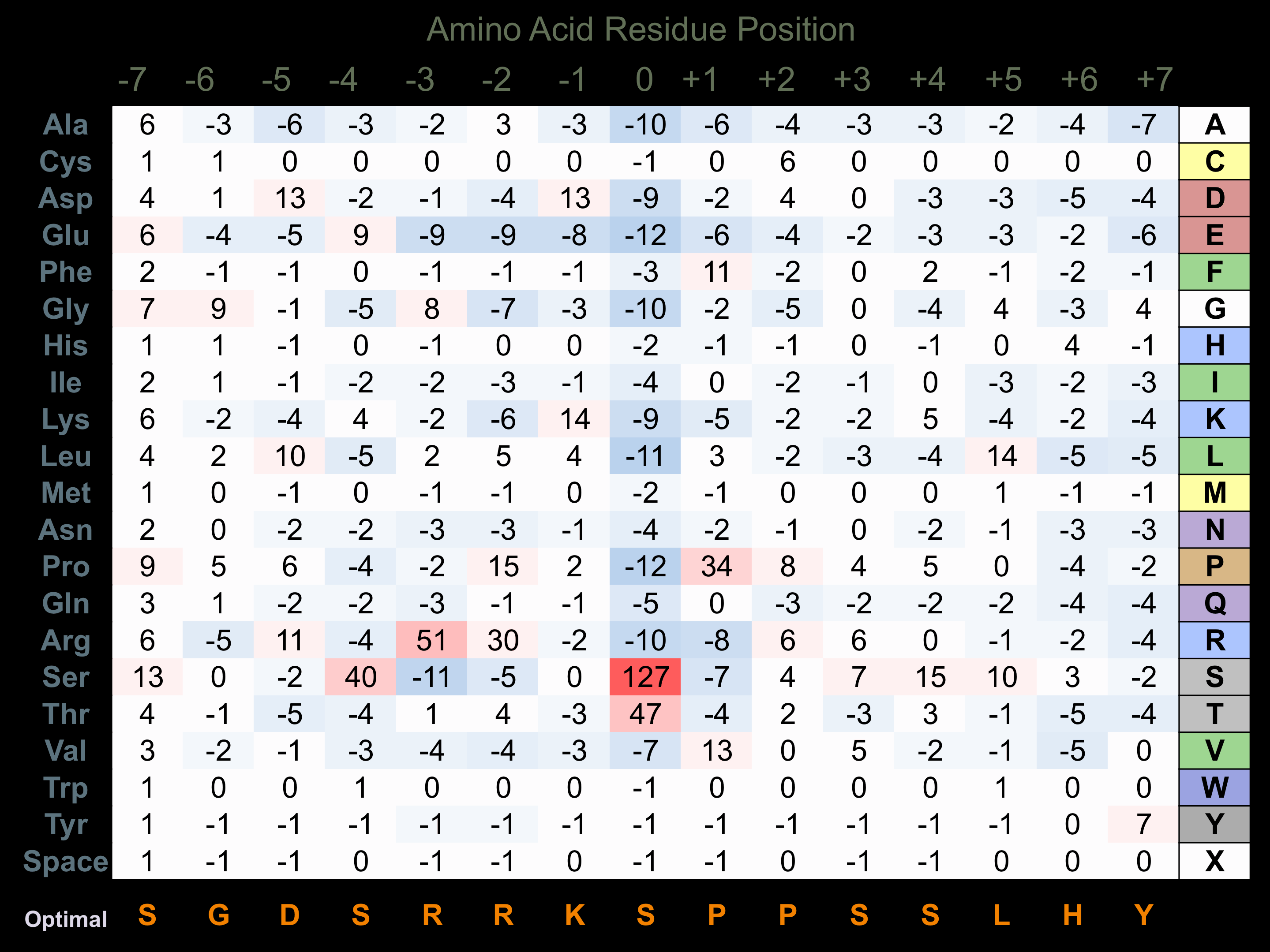
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 24721 diverse cancer specimens. This rate is only -20 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.28 % in 603 endometrium cancers tested; 0.25 % in 1259 large intestine cancers tested; 0.18 % in 273 cervix cancers tested; 0.17 % in 864 skin cancers tested; 0.17 % in 1276 kidney cancers tested; 0.09 % in 833 ovary cancers tested; 0.08 % in 575 stomach cancers tested; 0.07 % in 710 oesophagus cancers tested; 0.07 % in 1437 pancreas cancers tested; 0.06 % in 1634 lung cancers tested; 0.05 % in 1361 breast cancers tested; 0.04 % in 548 urinary tract cancers tested; 0.04 % in 2081 central nervous system cancers tested; 0.03 % in 942 upper aerodigestive tract cancers tested; 0.03 % in 881 prostate cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,005 cancer specimens
Comments:
Only 3 deletions, 1 insertion, and no complex mutations are noted on the COSMIC website.

