Nomenclature
Short Name:
SuRTK106
Full Name:
Tyrosine protein-kinase STYK1
Alias:
- DKFZP761P1010
- EC 2.7.10.2
- NOK
- NPAK
- STYK1
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
TK-Unique
SubFamily:
NA
Structure
Mol. Mass (Da):
47547
# Amino Acids:
422
# mRNA Isoforms:
1
mRNA Isoforms:
47,577 Da (422 AA; Q6J9G0)
4D Structure:
NA
1D Structure:
Subfamily Alignment
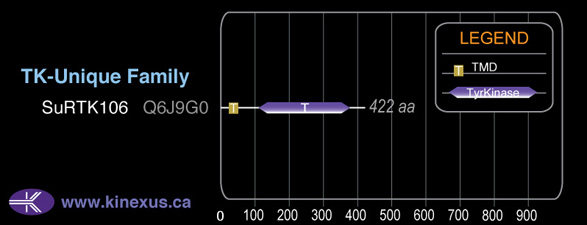
Domain Distribution:
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S59, S91.
Threonine phosphorylated:
T90, T224.
Tyrosine phosphorylated:
Y221.
Ubiquitinated:
K105.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 24
24
589
29
859
 0.6
0.6
14
13
23
 1.4
1.4
36
8
28
 9
9
235
85
309
 21
21
537
25
463
 0.8
0.8
20
74
19
 4
4
101
27
35
 13
13
338
33
426
 15
15
387
17
314
 0.7
0.7
17
61
23
 0.9
0.9
23
29
28
 17
17
436
142
384
 0.6
0.6
15
34
20
 0.8
0.8
19
9
21
 1
1
25
20
34
 0.6
0.6
15
14
14
 4
4
89
94
65
 0.9
0.9
22
16
23
 0.6
0.6
16
77
19
 12
12
303
109
335
 1.2
1.2
31
21
38
 0.9
0.9
23
23
18
 2
2
54
14
40
 0.7
0.7
17
15
20
 1
1
24
18
23
 11
11
279
51
348
 0.7
0.7
18
34
17
 2
2
56
19
44
 1
1
26
19
31
 4
4
102
28
99
 10
10
258
24
253
 100
100
2505
36
6370
 3
3
80
54
331
 20
20
502
52
432
 4
4
100
35
216
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
96 93.3
93.3
95.5
94 -
-
-
77 -
-
-
- 80.3
80.3
87.9
82 -
-
-
- 76.9
76.9
85.5
78 77.6
77.6
85.5
79 -
-
-
- 33.1
33.1
38.3
- 55.7
55.7
71.9
57 21.6
21.6
32.8
52 35.3
35.3
50.7
46 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
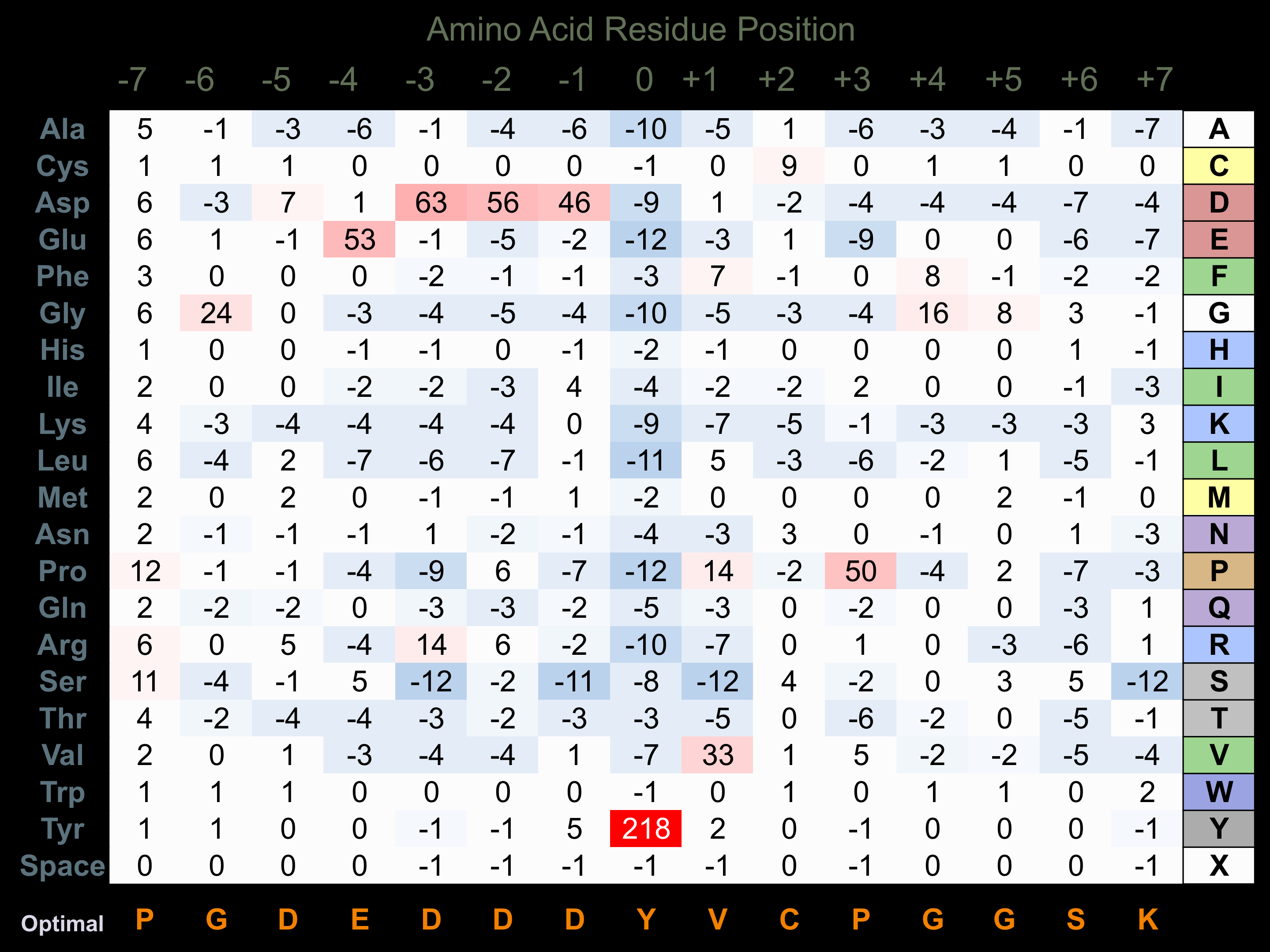
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Neurological disorder
Specific Diseases (Non-cancerous):
Chronic inflammatory demyelinating polyneuritis
Comments:
Chronic Inflammatory Demyelinating Polyneuritis is a neuronal disorder which is associated with the gene SuRTK106.
Comments:
SuRTK106 appears to be an oncoprotein (OP). With over-expression, SuRTK106 can induce tumour cell invasion or metastasis. A hypothetical mechanism is that SuRTK106 activates the MAP kinase and phosphatidyl 3’-kinase pathways.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Breast epithelial carcinomas (%CFC= -64, p<0.011); Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= +322, p<0.004); Colon mucosal cell adenomas (%CFC= -62, p<0.0001); Oral squamous cell carcinomas (OSCC) (%CFC= +122, p<0.0008); and Ovary adenocarcinomas (%CFC= +143, p<0.0002). The COSMIC website notes an up-regulated expression score for SuRTK106 in diverse human cancers of 398, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 6 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.1 % in 25653 diverse cancer specimens. This rate is only 31 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.82 % in 805 skin cancers tested; 0.37 % in 1093 large intestine cancers tested; 0.17 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: L316P (3).
Comments:
Only 1 complex mutation, and no deletions or insertions are noted on the COSMC website.