Nomenclature
Short Name:
TAK1
Full Name:
Mitogen-activated protein kinase kinase kinase 7
Alias:
- EC 2.7.11.25
- M3K7
- MAP3K7
- TGFa
- TGF-beta-activated kinase 1
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
Family:
MLK
SubFamily:
TAK1
Specific Links
Structure
Mol. Mass (Da):
67,196
# Amino Acids:
606
# mRNA Isoforms:
4
mRNA Isoforms:
67,196 Da (606 AA; O43318); 64,230 Da (579 AA; O43318-2); 56,706 Da (518 AA; O43318-3); 53,740 Da (491 AA; O43318-4)
4D Structure:
Binds both upstream activators and downstream substrates in multimolecular complexes. Interacts with TAB1/MAP3K7IP1 and TAB2/MAP3K7IP2. Identified in the TRIKA2 complex composed of MAP3K7, TAB1/MAP3K7IP1 and TAB2/MAP3K7IP2. Interacts with PPM1L. Interaction with PP2A and PPP6C leads to its repressed activity. Interacts with TRAF6 and TAB1/MAP3K7IP1; during IL-1 signaling. Interacts with TAOK1 and TAOK2; interaction with TAOK2 interferes with MAP3K7 interaction with IKKA, thus preventing NF-kappa-B activation. Interacts with WDR34 (via WD domains). Interacts with RBCK1. Interacts with CYLD.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 36 | 299 | Pkinase |
| 528 | 593 | Coiled-coil |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K386, K573.
Serine phosphorylated:
S192-, S367, S389, S393, S406, S417, S439, S454, S455, S458, S459.
Threonine phosphorylated:
T178+, T184+, T187+, T333, T415, T444.
Tyrosine phosphorylated:
Y405, Y584, Y585.
Ubiquitinated:
K72, K158, K209, K346, K351, K354, K590.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 87
87
1023
55
1044
 6
6
71
25
61
 19
19
221
19
103
 37
37
442
176
515
 79
79
928
47
705
 7
7
88
152
38
 16
16
189
55
390
 100
100
1182
74
2527
 56
56
664
31
475
 10
10
122
148
83
 10
10
124
56
94
 69
69
811
279
667
 11
11
132
63
75
 9
9
104
18
52
 11
11
129
44
116
 9
9
110
29
66
 13
13
159
274
140
 16
16
190
31
135
 15
15
179
160
102
 57
57
675
215
660
 16
16
195
40
148
 21
21
247
47
222
 19
19
227
23
137
 17
17
197
32
149
 21
21
245
40
260
 54
54
640
104
651
 15
15
173
66
136
 12
12
137
31
85
 18
18
209
32
151
 11
11
128
56
111
 61
61
716
36
794
 52
52
615
72
706
 12
12
142
105
383
 81
81
960
104
694
 10
10
118
61
117
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 74.4
74.4
75.7
100 99.5
99.5
99.5
100 -
-
-
100 -
-
-
- -
-
-
100 -
-
-
- 95
95
95
99 99
99
99.3
99 -
-
-
- 91.4
91.4
93.7
- 91.8
91.8
93.7
92 22
22
35.7
96 70.6
70.6
77.9
- -
-
-
- 34.5
34.5
52.2
35 41.4
41.4
58.8
- -
-
-
25 39.9
39.9
51.5
- -
-
-
- -
-
-
- -
-
-
- -
-
-
36 -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | TAB1 - Q15750 |
| 2 | TAB3 - Q8N5C8 |
| 3 | STAT3 - P40763 |
| 4 | SMAD6 - O43541 |
| 5 | CDC37 - Q16543 |
| 6 | NLK - Q9UBE8 |
| 7 | BCL10 - O95999 |
| 8 | PPM1B - O75688 |
| 9 | NAIP - Q13075 |
| 10 | IL17RD - Q8NFM7 |
| 11 | NRIP1 - P48552 |
| 12 | MAP4K4 - O95819 |
| 13 | MAP2K4 - P45985 |
| 14 | PPM1L - Q5SGD2 |
| 15 | RELA - Q04206 |
Regulation
Activation:
Activated by phosphorylation at Thr-178, Thr-184, Thr-187 and Ser-192. Activated by 'Lys-63'-linked polyubiquitination.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| DSCR1 | P53805 | S149 | QTLHIGSSHLAPPNP | |
| DSCR1 | P53805 | S191 | YDLLYAISKLGPGEK | |
| HDAC4 | P56524 | S246 | FPLRKTASEPNLKLR | |
| HDAC5 | Q9UQL6 | S259 | FPLRKTASEPNLKVR | |
| HDAC7 | Q8WUI4 | S155 | FPLRKTVSEPNLKLR | |
| Ksr1 (KSR) | Q8IVT5 | S404 | TRLRRTESVPSDINN | |
| PTPN3 | P26045 | S359 | PAMRRSLSVEHLETK | |
| TAK1 (MAP3K7) | O43318 | S192 | HMTNNKGSAAWMAPE | - |
| TAK1 (MAP3K7) | O43318 | T184 | GTACDIQTHMTNNKG | + |
| TAK1 (MAP3K7) | O43318 | T187 | CDIQTHMTNNKGSAA | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
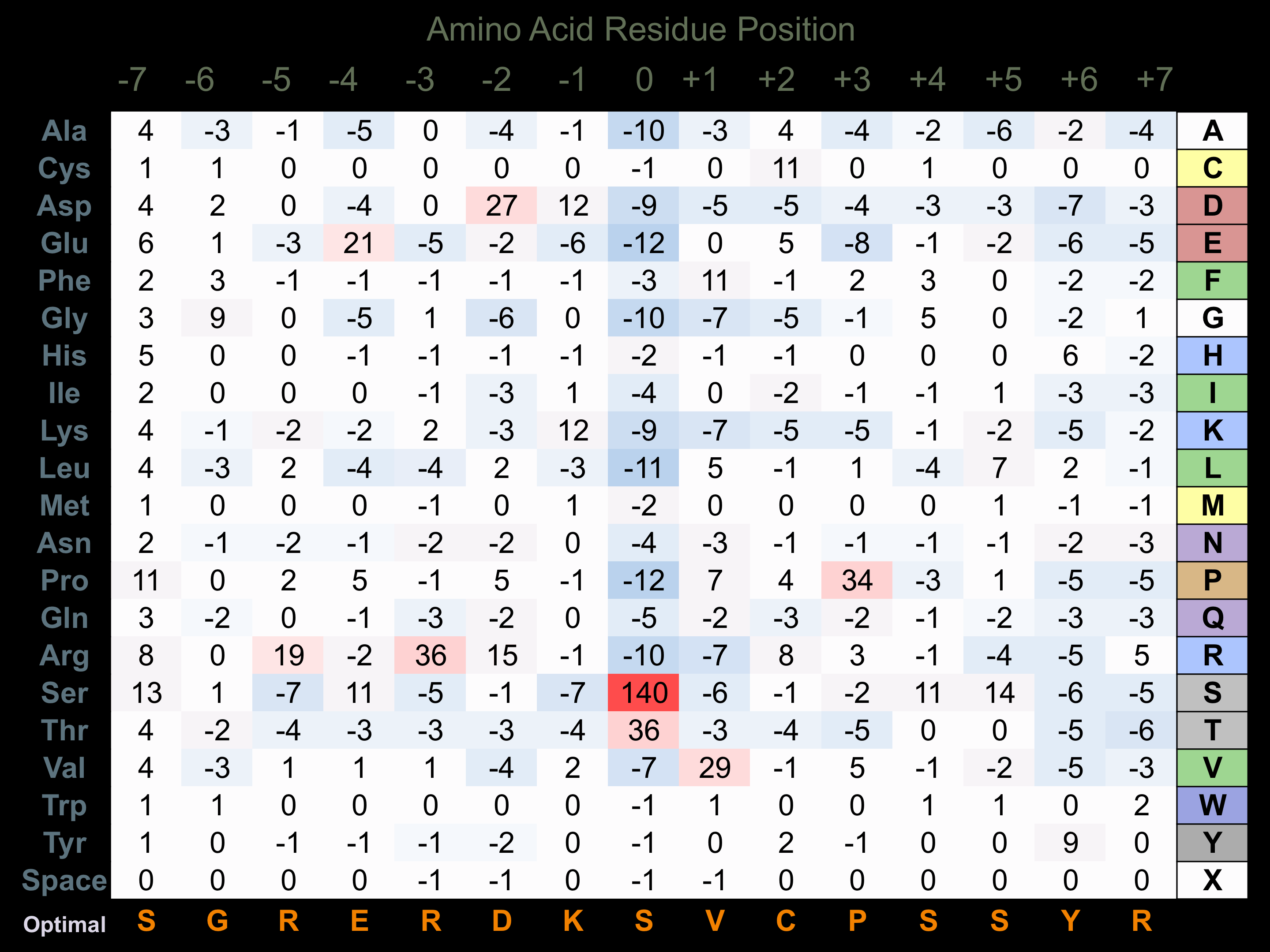
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Skin disorder
Specific Diseases (Non-cancerous):
Hypohidrotic ectodermal dysplasia (EDA)
Comments:
TAK1 mediates signal transduction induced by TGF beta and morphogenetic protein, functions in transcription regulation and apoptosis. It regulates hepatic cell survival and carcinogenesis. A K63W substitution in TAK1 is associated with loss of kinase activity.
Comments:
TAK1 plays a role in tumour initiation, progression, and metastasis as a tumour inducer or tumour suppressor.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +45, p<0.0001); Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= +65, p<0.1); and Prostate cancer (%CFC= +66, p<0.005). The COSMIC website notes an up-regulated expression score for TAK1 in diverse human cancers of 351, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 302 for this protein kinase in human cancers was 5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. A K63W substitution in TAK1 can result in reduction in its phoshotransferase activity.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25474 diverse cancer specimens. This rate is very similar (+ 1% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.45 % in 1093 large intestine cancers tested; 0.27 % in 602 endometrium cancers tested; 0.14 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R83C (4).
Comments:
Only 4 deletions, 1 insertion, and 1 complex mutation are noted on the COSMIC website.

