Nomenclature
Short Name:
TBCK
Full Name:
TBC domain-containing protein kinase-like protein
Alias:
- BAB85045
- HSPC302
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
TBCK
SubFamily:
NA
Structure
Mol. Mass (Da):
100,680
# Amino Acids:
893
# mRNA Isoforms:
3
mRNA Isoforms:
100,679 Da (893 AA; Q8TEA7); 96,443 Da (854 AA; Q8TEA7-2); 93,725 Da (830 AA; Q8TEA7-3)
4D Structure:
NA
1D Structure:
Subfamily Alignment
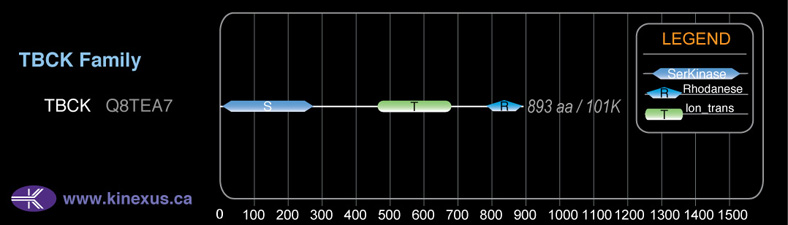
Domain Distribution:
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S118, S890.
Threonine phosphorylated:
T169.
Tyrosine phosphorylated:
Y153, Y732.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 85
85
1249
9
943
 3
3
51
4
46
 7
7
100
23
71
 29
29
426
61
919
 70
70
1021
13
760
 11
11
160
9
65
 8
8
117
13
52
 49
49
719
36
1849
 2
2
25
3
1
 18
18
264
54
175
 5
5
80
30
54
 24
24
358
47
515
 8
8
112
23
63
 6
6
84
3
6
 4
4
61
7
50
 14
14
211
5
26
 4
4
53
170
56
 5
5
74
26
54
 4
4
62
44
33
 81
81
1189
31
741
 8
8
124
30
90
 8
8
122
30
89
 11
11
160
23
92
 9
9
127
26
91
 7
7
97
30
80
 40
40
588
49
674
 6
6
83
26
56
 6
6
95
26
68
 12
12
176
26
127
 6
6
85
14
55
 100
100
1468
12
24
 5
5
72
11
83
 17
17
245
43
390
 80
80
1178
26
813
 51
51
744
22
589
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.5
99.5
99.7
100 -
-
-
97 -
-
-
96.5 -
-
-
- 96.7
96.7
98.8
97 -
-
-
- 81
81
83.5
95 90.5
90.5
94.2
94 -
-
-
- 90.3
90.3
94.5
- 87.1
87.1
92.8
87 81.5
81.5
90
81.5 76.7
76.7
87.6
77 -
-
-
- 43.6
43.6
58.5
48 47.5
47.5
62.7
- -
-
-
34 55.2
55.2
70.8
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 20
20
32.3
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
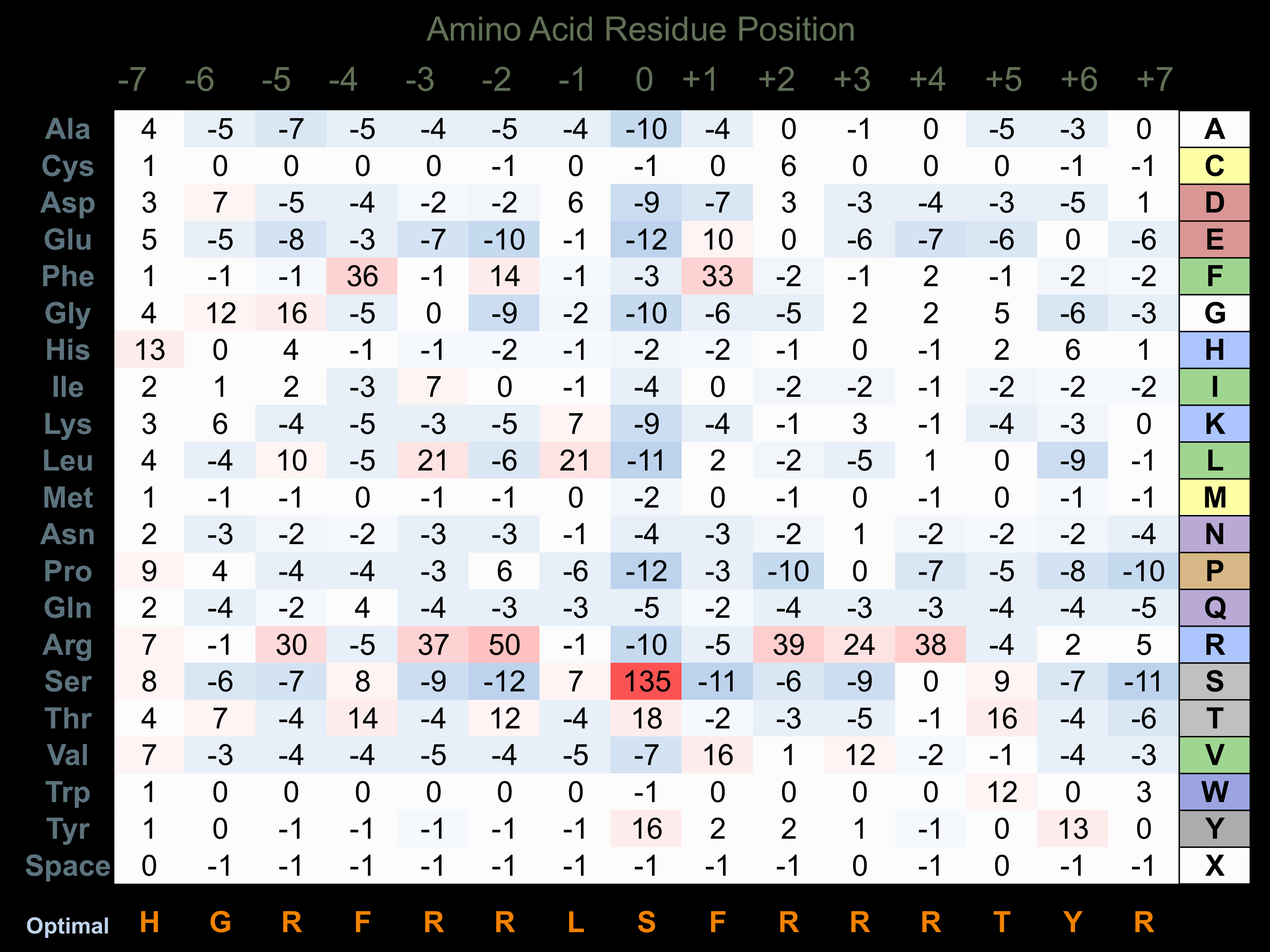
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for TBCK in diverse human cancers of 284, which is 0.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 132 for this protein kinase in human cancers was 2.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24727 diverse cancer specimens. This rate is only -4 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.36 % in 1270 large intestine cancers tested; 0.23 % in 864 skin cancers tested; 0.23 % in 589 stomach cancers tested; 0.22 % in 603 endometrium cancers tested; 0.14 % in 1634 lung cancers tested; 0.1 % in 548 urinary tract cancers tested; 0.1 % in 1512 liver cancers tested; 0.09 % in 127 biliary tract cancers tested; 0.08 % in 1459 pancreas cancers tested; 0.06 % in 382 soft tissue cancers tested; 0.05 % in 881 prostate cancers tested; 0.04 % in 833 ovary cancers tested; 0.04 % in 273 cervix cancers tested; 0.03 % in 710 oesophagus cancers tested; 0.03 % in 1316 breast cancers tested; 0.03 % in 1276 kidney cancers tested; 0.02 % in 942 upper aerodigestive tract cancers tested; 0.02 % in 558 thyroid cancers tested; 0.01 % in 2082 central nervous system cancers tested; 0.01 % in 2009 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R41C (4); Q504H (4).
Comments:
Eleven of the deletions mutants were at N457 located in the Rab-GAP domain.

