Nomenclature
Short Name:
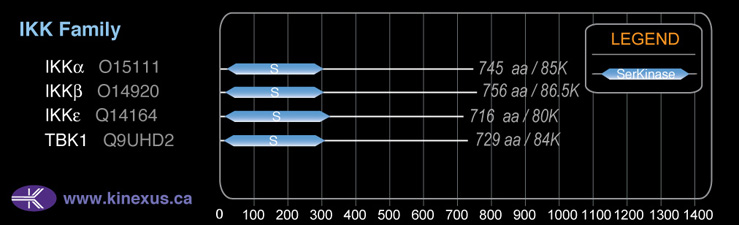
TBK1
Full Name:
TANK binding kinase TBK1
Alias:
- EC 2.7.11.1
- FLJ11330
- TANK-binding kinase 1
- Kinase TBK1
- NAK
- NF-KB-activating kinase NAK
- TANK binding kinase TBK1
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
IKK
SubFamily:
NA
Structure
Mol. Mass (Da):
83,642
# Amino Acids:
729
# mRNA Isoforms:
1
mRNA Isoforms:
83,642 Da (729 AA; Q9UHD2)
4D Structure:
Interacts with TIRAP, TANK and TRAF2. Part of a ternary complex consisting of TANK, TRAF2 and TBK1. Interacts with AZI2. Interacts with SIKE1. Interacts with TICAM1/TRIF, IRF3 and DDX58/RIG-I, interactions are disrupted by the interaction between TBK1 and SIKE1. Interacts with CYLD. Interacts with HCV NS3, a hepatitis C virus protein and with BDV P protein, a Borna disease virus protein
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 9 | 311 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S151, S172+, S247, S511, S716.
Threonine phosphorylated:
T503, T664.
Tyrosine phosphorylated:
Y153, Y174+, Y179, Y354, Y577.
Ubiquitinated:
K30, K69, K137, K154, K241, K291, K344, K372, K396, K401, K504, K584, K661.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 51
51
929
21
867
 4
4
67
12
50
 10
10
184
1
0
 22
22
398
70
490
 41
41
738
24
726
 7
7
118
46
64
 5
5
85
28
82
 40
40
718
22
1062
 30
30
534
13
612
 7
7
122
36
46
 8
8
137
15
96
 45
45
807
83
688
 7
7
123
12
22
 6
6
101
9
83
 4
4
68
12
54
 6
6
107
12
92
 3
3
58
97
64
 3
3
52
9
60
 10
10
175
40
126
 36
36
644
82
596
 6
6
101
11
75
 7
7
135
13
116
 10
10
176
2
79
 7
7
132
9
179
 2
2
45
11
50
 34
34
608
45
645
 6
6
109
18
65
 3
3
52
9
53
 4
4
79
9
73
 11
11
191
14
80
 67
67
1217
18
677
 28
28
515
26
678
 9
9
160
68
449
 53
53
965
52
800
 100
100
1809
35
2794
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.6
99.6
99.9
100 99
99
99.9
99 -
-
-
97 -
-
-
- 96.7
96.7
98.9
97 -
-
-
- 94.1
94.1
97.7
94 22.1
22.1
42.5
94.5 -
-
-
- 88.2
88.2
94.7
- 20.2
20.2
39.9
87 73.4
73.4
86.8
76 21
21
41.8
72 -
-
-
- 20.5
20.5
39.5
41.5 38
38
57.8
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | TANK - Q92844 |
| 2 | IRF3 - Q14653 |
| 3 | RELA - Q04206 |
| 4 | GAPDH - P04406 |
| 5 | TICAM1 - Q8IUC6 |
| 6 | NCK1 - P16333 |
| 7 | CYLD - Q9NQC7 |
| 8 | NFKBIA - P25963 |
| 9 | REL - Q04864 |
| 10 | NFKB1 - P19838 |
| 11 | RUVBL2 - Q9Y230 |
| 12 | RPS5 - P46782 |
| 13 | HNRNPM - P52272 |
| 14 | SKP1 - P63208 |
| 15 | NONO - Q15233 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| IKKb (IKBKINASE) | O14920 | S177 | AKELDQGSLCTSFVG | + |
| InsR | P06213 | S1033 | LRELGQGSFGMVYEG | |
| IRF3 | Q14653 | S173 | PCPQPLRSPSLDNPT | + |
| IRF3 | Q14653 | S175 | PQPLRSPSLDNPTPF | |
| IRF3 | Q14653 | S385 | MARVGGASSLENTVD | + |
| IRF3 | Q14653 | S386 | ARVGGASSLENTVDL | + |
| IRF3 | Q14653 | S396 | NTVDLHISNSHPLSL | + |
| IRF3 | Q14653 | S398 | VDLHISNSHPLSLTS | + |
| IRF3 | Q14653 | S402 | ISNSHPLSLTSDQYK | + |
| IRF3 | Q14653 | S405 | SHPLSLTSDQYKAYL | + |
| IRF3 | Q14653 | T404 | NSHPLSLTSDQYKAY | + |
| IRF7 | Q92985 | S471 | GTQREGVSSLDSSSL | - |
| IRF7 | Q92985 | S472 | TQREGVSSLDSSSLS | - |
| IRF7 | Q92985 | S477 | VSSLDSSSLSLCLSS | + |
| IRF7 | Q92985 | S479 | SLDSSSLSLCLSSAN | + |
| NFkB-p65 | Q04206 | S536 | SGDEDFSSIADMDFS |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
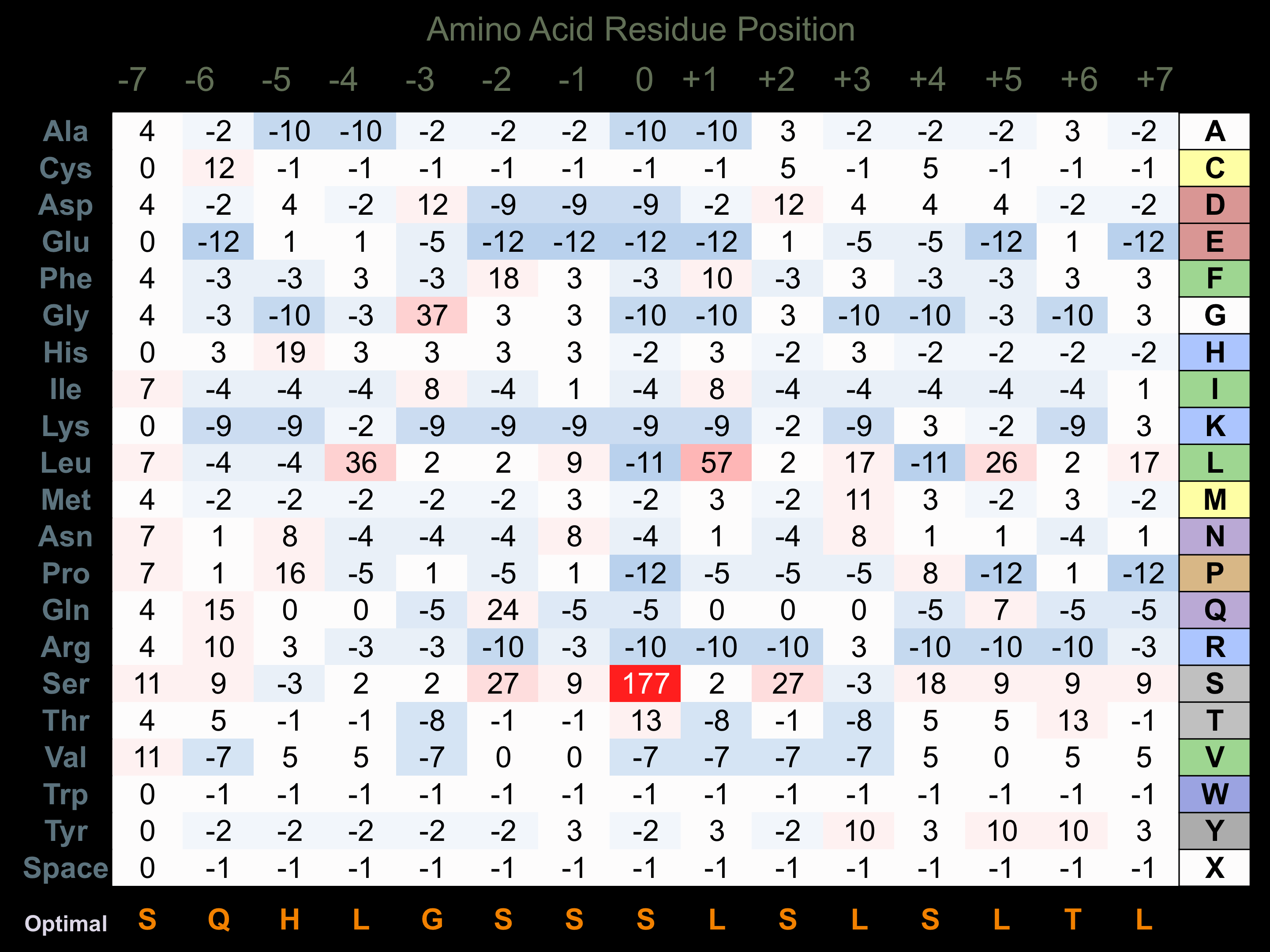
Matrix Type:
Experimentally derived from alignment of 22 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Infectious disease, and eye disorder
Specific Diseases (Non-cancerous):
Herpes simplex; Borna disease; Herpes simplex Encephalitis; Herpes simplex Encephalitis 1; Glaucoma 1, open angle, P (GLC1P)
Comments:
TBK1 has been linked to Herpes Simplex, which is a rare infectious reproductive disease characterized by cold sores affecting the face or mouth. Herpes Simplex has been related to the encephalitis and genital herpes diseases. Herpes Simplex Encephalitis has been related to encephalitis and Herpes Simplex diseases. Herpes Simplex Encephalitis can affect the brain, temporal lobe, and testes. Herpes Simplex Encephalitis 1 is a rare disorder that can affect the skin and brain. Glaucoma 1, open angle, P (GLC1P) can affect the optic nerve, reducing the range of the visual field.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +81, p<0.0009); Breast epithelial carcinomas (%CFC= -56, p<0.102); Oral squamous cell carcinomas (OSCC) (%CFC= +69, p<(0.0003); and T-cell prolymphocytic leukemia (%CFC= -59, p<0.0009).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. Ubiquitination of TBK1 can be decreased with a K30R, or K401R mutation. TBK1 phosphotransferase activity can be inhibited with a K38A, D135N, S172A, or Y325E (through inhibited phosphorylation) mutation. The phosphotransferase activity canalso be decreased with a D33A (with decreased phosphorylation), S172E, L316E (does not affect phosphorylation), E355R, R357A, E448R (via decreased phosphorylation), R547D (via decreased phosphorylation), Y577A, E580A, I582A, or K589D (via decreased phosphorylation) mutations. Dimerization of TBK1 can be inhibited with the E355R + R357A (in conjunction), or E355R + E448R, or H459E + I466E + F470E (which will also decrease kinase phosphotransferase activity, yet not affect phosphorylation levels), or R547D mutations. Phosphorylation, dimerization, and ubiquitination can be inhibited with the K401R + K30R (in conjunction) mutations. TBK1 interaction with TANK can be decreased with a TBK1 mutation, M690A. Interaction can be fully inhibited with a L693A, or N708A mutation. Interaction with TANK as well as TBKBP1 can be decreased with a K694E mutation, and a L704A mutation will further decrease interaction with AZI2. Interaction with TANK can be almost fully abrogated with a L711A mutation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 24865 diverse cancer specimens. This rate is only -20 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: E109K (10).
Comments:
Only 3 deletions, and no insertions or complex mutations are noted on the COSMIC website.

