Nomenclature
Short Name:
TESK1
Full Name:
Dual-specificity testis-specific protein kinase 1
Alias:
- TES1
- Testicular protein kinase 1
- Testis specific kinase-1
- Testis-specific kinase 1
- Testis-specific kinase-1
- Testis-specific protein kinase 1
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
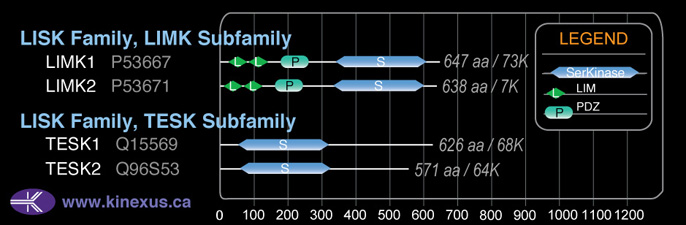
Family:
LISK
SubFamily:
TESK
Structure
Mol. Mass (Da):
67684
# Amino Acids:
626
# mRNA Isoforms:
1
mRNA Isoforms:
67,684 Da (626 AA; Q15569)
4D Structure:
Interacts with SPRY4.
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 57 | 315 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K391, K393, K400.
Serine phosphorylated:
S69, S220-, S357, S437+, S440+, S441, S553.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 39
39
725
16
842
 12
12
234
10
192
 72
72
1360
11
1283
 29
29
539
60
756
 39
39
741
14
699
 7
7
128
46
199
 21
21
386
19
625
 60
60
1122
34
1452
 16
16
299
10
261
 16
16
301
53
458
 23
23
441
22
607
 34
34
632
115
641
 34
34
632
22
982
 3
3
60
7
53
 29
29
539
20
692
 11
11
207
8
129
 18
18
335
110
660
 43
43
806
16
1231
 26
26
490
60
826
 33
33
618
56
613
 31
31
589
18
642
 35
35
654
22
945
 28
28
535
20
678
 65
65
1230
18
1923
 48
48
900
18
1394
 100
100
1881
42
3052
 23
23
429
25
707
 41
41
762
17
1361
 37
37
687
18
856
 5
5
87
14
70
 55
55
1040
18
714
 31
31
578
21
631
 22
22
411
51
601
 38
38
712
31
610
 14
14
267
22
217
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 77.3
77.3
77.4
100 98.9
98.9
99.2
99 -
-
-
95 -
-
-
95 43.3
43.3
53.8
93 -
-
-
- 93.1
93.1
95.2
93 92.2
92.2
94.3
92.5 -
-
-
- 40.9
40.9
50.5
- 40.9
40.9
53.2
66 -
-
-
- 46.3
46.3
59.6
66 -
-
-
- 22
22
31.3
- 24.9
24.9
34.4
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Phosphorylation of Ser-220 and Ser-440 increases phosphotransferase activity.
Inhibition:
Binding of 14-3-3 beta inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
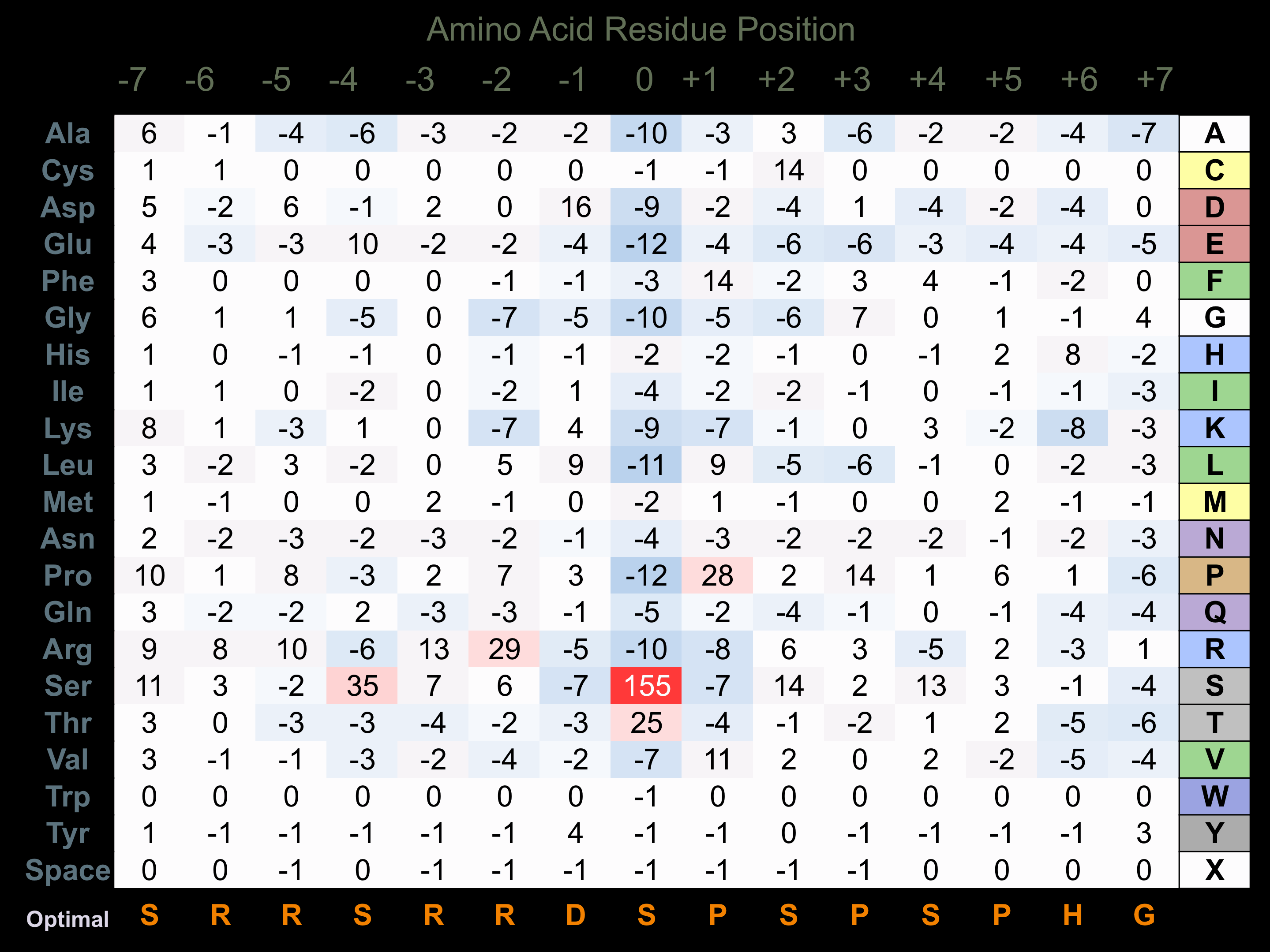
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Breast epithelial cell carcinomas (%CFC= +83, p<0.004); Classical Hodgkin lymphomas (%CFC= +63, p<0.004); Large B-cell lymphomas (%CFC= +157, p<0.002); Ovary adenocarcinomas (%CFC= +103, p<0.034); Pituitary adenomas (ACTH-secreting) (%CFC= -66); Pituitary adenomas (aldosterone-secreting) (%CFC= +89, p<0.038); and Uterine leiomyomas (%CFC= +125, p<0.091). The COSMIC website notes an up-regulated expression score for TESK1 in diverse human cancers of 512, which is 1.1-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 70 for this protein kinase in human cancers was 1.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 25034 diverse cancer specimens. This rate is only -29 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.41 % in 589 stomach cancers tested; 0.23 % in 1296 large intestine cancers tested; 0.16 % in 603 endometrium cancers tested; 0.13 % in 127 biliary tract cancers tested; 0.12 % in 1823 lung cancers tested; 0.11 % in 864 skin cancers tested; 0.08 % in 833 ovary cancers tested; 0.06 % in 273 cervix cancers tested; 0.05 % in 2082 central nervous system cancers tested.
Frequency of Mutated Sites:
None > 2 in 20,318 cancer specimens
Comments:
Only 2 deletions, and no insertions or complex mutations are noted on the COSMIC website.

