Nomenclature
Short Name:
TSSK4
Full Name:
Testis-specific serine-threonine protein kinase 4
Alias:
- EC 2.7.11.1
- C14orf20
- STK22E
- TSK4
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
TSSK
SubFamily:
NA
Structure
Mol. Mass (Da):
37,454
# Amino Acids:
328
# mRNA Isoforms:
3
mRNA Isoforms:
38,439 Da (338 AA; Q6SA08-2); 37,454 Da (328 AA; Q6SA08); 29,034 Da (252 AA; Q6SA08-3)
4D Structure:
NA
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 25 | 293 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S165, S264.
Threonine phosphorylated:
T197+, T251, T256, T283.
Ubiquitinated:
K281.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 39
39
816
6
936
 6
6
122
6
187
 -
-
-
-
-
 11
11
239
33
1009
 16
16
340
12
294
 0.4
0.4
8
9
6
 0.2
0.2
5
13
4
 13
13
270
9
527
 0.1
0.1
3
3
1
 3
3
66
33
152
 6
6
129
9
248
 27
27
566
15
559
 15
15
318
2
28
 16
16
340
5
688
 23
23
480
9
1111
 10
10
205
7
340
 1
1
20
88
109
 11
11
236
5
367
 9
9
192
23
535
 22
22
452
33
583
 10
10
216
9
425
 0.2
0.2
4
7
4
 -
-
-
-
-
 100
100
2082
7
2037
 3
3
58
9
135
 30
30
622
21
576
 12
12
255
5
347
 15
15
322
5
443
 11
11
238
5
410
 7
7
141
14
69
 25
25
526
12
52
 5
5
97
7
54
 0.1
0.1
3
12
0
 36
36
746
26
650
 23
23
483
22
404
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 98.8
98.8
99.1
99 93.6
93.6
96.3
94 -
-
-
89 -
-
-
- 87.2
87.2
93.9
87 -
-
-
- 85.7
85.7
92.7
86 42.7
42.7
46.3
89 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 38.3
38.3
56.7
42 42.7
42.7
60.7
- -
-
-
37 42.7
42.7
61.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CREB1 - P16220 |
| 2 | SMURF2 - Q9HAU4 |
Regulation
Activation:
Activated by phosphorylation on Thr-197, potentially by autophosphorylation.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
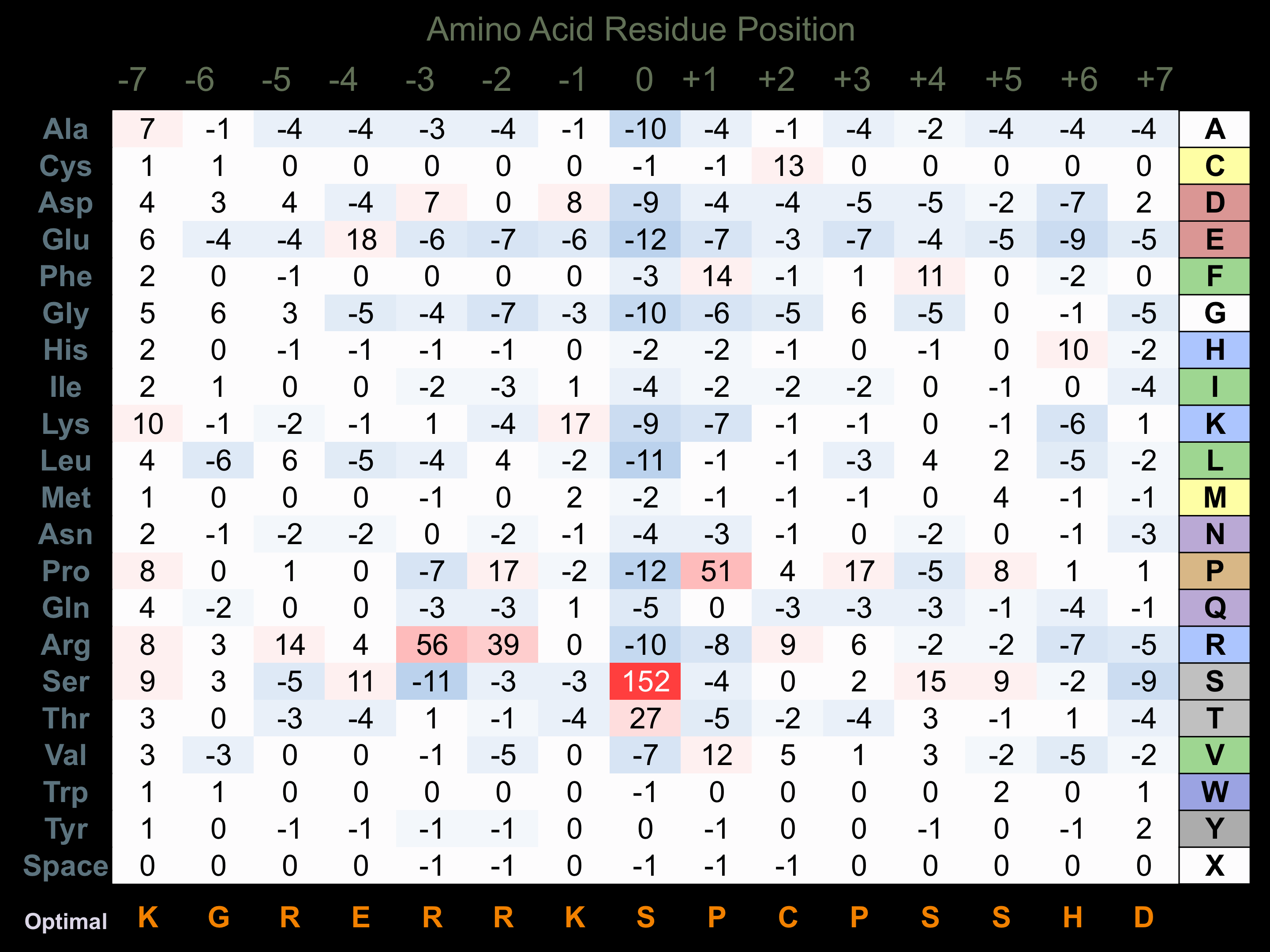
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Bone, and neuronal disorder
Specific Diseases (Non-cancerous):
Spinal stenosis; Spondylosis; Spondylolisthesis
Comments:
Spinal Stenosis can be characterized by the narrowing of the spine, neck pain, back numbness, weakness, cramping, arm pain, or leg pain. Spondylosis is the development of osteoarthritis in the joint between the neural foramina and the spinal vertebrae. Spondylolisthesis is often characterized by the displacement of the fifth lumbar vertebra.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= -68); and Brain oligodendrogliomas (%CFC= -76). The COSMIC website notes an up-regulated expression score for TSSK4 in diverse human cancers of 424, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 8 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 24658 diverse cancer specimens. This rate is only -39 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.24 % in 1259 large intestine cancers tested; 0.21 % in 864 skin cancers tested; 0.21 % in 589 stomach cancers tested; 0.15 % in 603 endometrium cancers tested; 0.11 % in 273 cervix cancers tested; 0.07 % in 1634 lung cancers tested; 0.05 % in 1276 kidney cancers tested.
Frequency of Mutated Sites:
None > 2 in 19,942 cancer specimens
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.

