Nomenclature
Short Name:
Trb1
Full Name:
Tribbles homolog 1
Alias:
- TRB-1
- SKIP1
- G-protein-coupled receptor induced protein 2
- GIG-2
- TRIB1
- C8FW
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
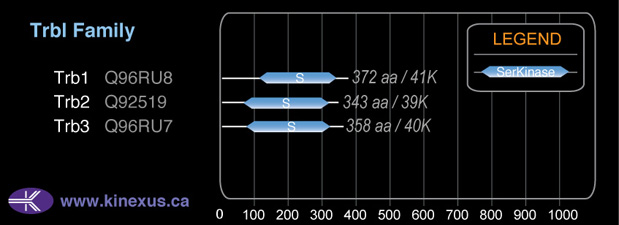
Family:
Trbl
SubFamily:
NA
Structure
Mol. Mass (Da):
41,009
# Amino Acids:
372
# mRNA Isoforms:
2
mRNA Isoforms:
41,009 Da (372 AA; Q96RU8); 23,473 Da (206 AA; Q96RU8-2)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 91 | 338 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S8.
Threonine phosphorylated:
T26, T229.
Ubiquitinated:
K233.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 81
81
1228
31
1246
 8
8
123
12
97
 5
5
78
3
96
 32
32
486
100
1347
 71
71
1085
29
936
 30
30
453
63
824
 19
19
292
39
482
 66
66
1009
27
1732
 53
53
808
10
694
 12
12
185
97
230
 32
32
488
20
626
 59
59
902
134
770
 12
12
180
14
100
 3
3
51
8
43
 8
8
127
17
110
 14
14
221
18
340
 27
27
408
231
554
 7
7
111
10
102
 18
18
271
81
296
 40
40
617
112
671
 4
4
59
16
40
 10
10
156
17
141
 17
17
255
12
174
 5
5
72
9
62
 8
8
120
14
112
 100
100
1525
65
1828
 10
10
157
17
98
 12
12
179
10
119
 7
7
103
10
133
 5
5
74
42
81
 54
54
823
18
585
 22
22
340
31
491
 24
24
362
80
721
 57
57
871
83
745
 42
42
634
48
2025
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.4
99.4
100
99.5 98.9
98.9
99.4
99 -
-
-
97 -
-
-
97 96.7
96.7
98.3
97 -
-
-
- 93.8
93.8
96.2
94 47
47
61
93.5 -
-
-
- 57.6
57.6
64.3
- 25.5
25.5
40
91 23.7
23.7
38.5
75.5 53.4
53.4
66.9
- -
-
-
- -
-
-
- 42.4
42.4
59.6
- -
-
-
- 46.2
46.2
60.2
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | ALOX12 - P18054 |
| 2 | MAP2K4 - P45985 |
| 3 | MAP2K1 - Q02750 |
| 4 | ATF4 - P18848 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
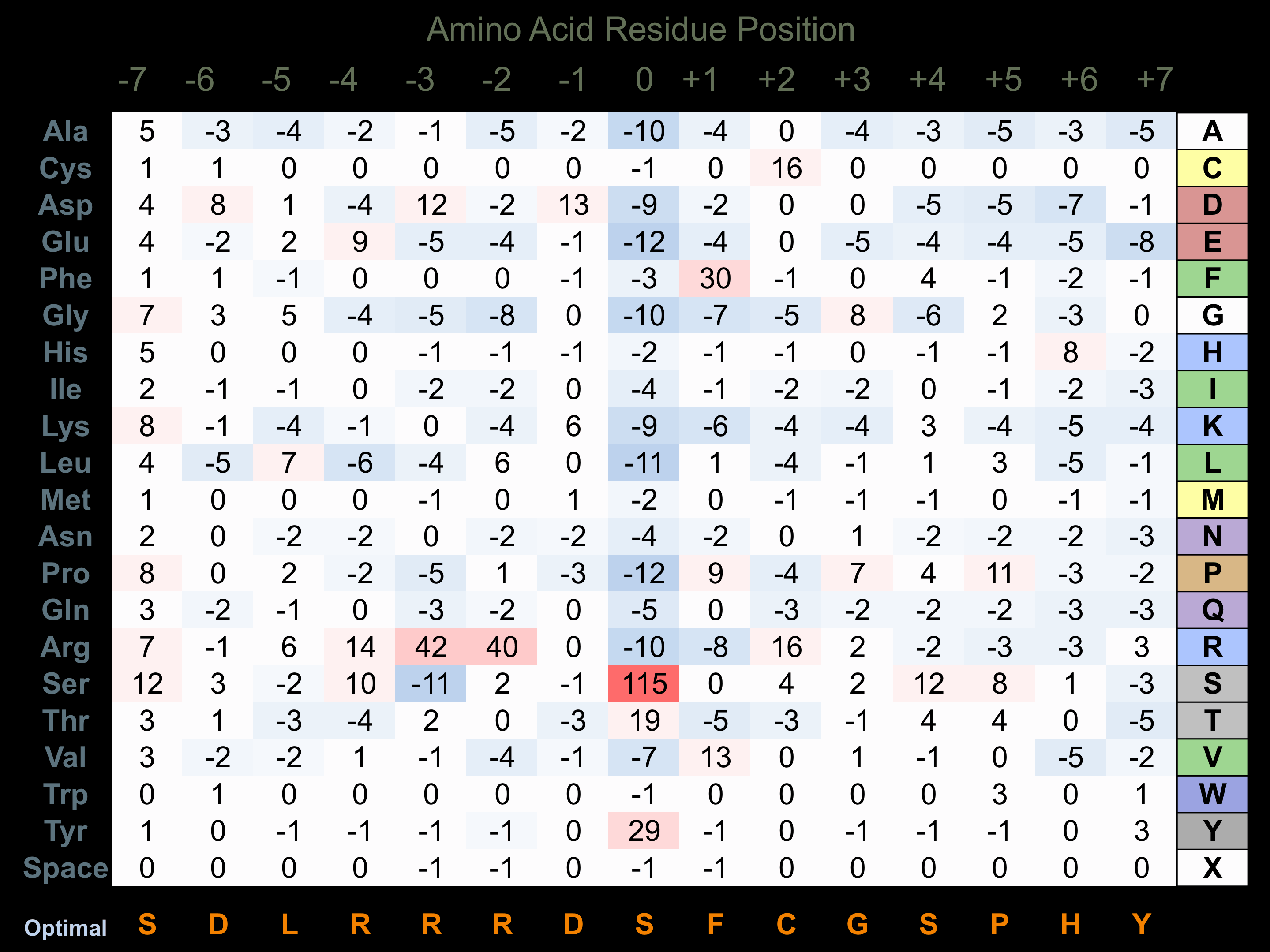
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Familial hypercholesterolemia (FH); Megakaryocytic leukemias (MKL)
Comments:
Trb1 was found overexpressed in HeLa cells, which contribute to repressed IL8 promoter. The gene was identified as cytogenetic hallmark of genomic amplification in 1% of karyotypically abnormal acute myeloid leukemias. The protein was also shown to support prostate tumorigenesis and tumour-propagating cell survival by regulation of endoplasmic reticulum chaperone expression.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 24433 diverse cancer specimens. This rate is very similar (+ 10% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.4 % in 603 endometrium cancers tested; 0.37 % in 1229 large intestine cancers tested; 0.31 % in 864 skin cancers tested; 0.21 % in 382 soft tissue cancers tested; 0.15 % in 710 oesophagus cancers tested; 0.15 % in 555 stomach cancers tested; 0.15 % in 1437 pancreas cancers tested; 0.11 % in 238 bone cancers tested; 0.08 % in 1608 lung cancers tested; 0.08 % in 1289 breast cancers tested; 0.06 % in 881 prostate cancers tested; 0.06 % in 1253 kidney cancers tested; 0.05 % in 558 thyroid cancers tested; 0.05 % in 548 urinary tract cancers tested; 0.04 % in 1512 liver cancers tested; 0.03 % in 807 ovary cancers tested; 0.01 % in 1982 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R297W (5); F317S (3).
Comments:
Only 2 deletions and 1 insertion, and no complex mutations are noted on the COSMIC website.

