Nomenclature
Short Name:
Trb2
Full Name:
Tribbles homolog 2
Alias:
- TRIB2
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
Trbl
SubFamily:
NA
Structure
Mol. Mass (Da):
38,801
# Amino Acids:
343
# mRNA Isoforms:
1
mRNA Isoforms:
38,801 Da (343 AA; Q92519)
4D Structure:
NA
1D Structure:
Subfamily Alignment
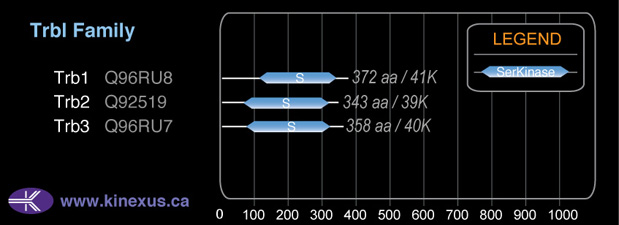
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 61 | 308 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S28, S29, S83, S133, S139, S299.
Threonine phosphorylated:
T143, T298.
Tyrosine phosphorylated:
Y14, Y134.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 21
21
921
29
800
 5
5
232
17
139
 6
6
254
4
311
 20
20
877
95
1431
 21
21
931
25
803
 15
15
669
86
1950
 8
8
353
35
594
 20
20
888
42
1091
 9
9
398
17
301
 4
4
192
89
156
 3
3
114
27
177
 19
19
814
188
663
 5
5
239
26
269
 7
7
295
15
170
 3
3
151
21
286
 6
6
249
16
225
 6
6
259
200
660
 4
4
185
15
300
 2
2
105
85
65
 12
12
529
109
534
 6
6
249
19
206
 9
9
399
23
368
 8
8
344
14
278
 5
5
236
15
171
 9
9
382
19
420
 26
26
1128
59
1998
 5
5
217
29
181
 4
4
193
15
241
 5
5
236
15
192
 4
4
174
28
134
 30
30
1316
24
685
 100
100
4397
36
8523
 29
29
1269
74
1741
 15
15
660
57
607
 3
3
133
35
94
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.1
99.1
99.1
100 91
91
92.7
- -
-
-
99 -
-
-
92 54.3
54.3
68.3
100 -
-
-
- 98.3
98.3
99.1
98 47.3
47.3
63.3
97 -
-
-
- 94.5
94.5
97.4
- 25.3
25.3
40
96 24.6
24.6
39.7
89.5 78.4
78.4
88.6
79 -
-
-
- -
-
-
44 42.9
42.9
62
- -
-
-
- 52.5
52.5
70
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
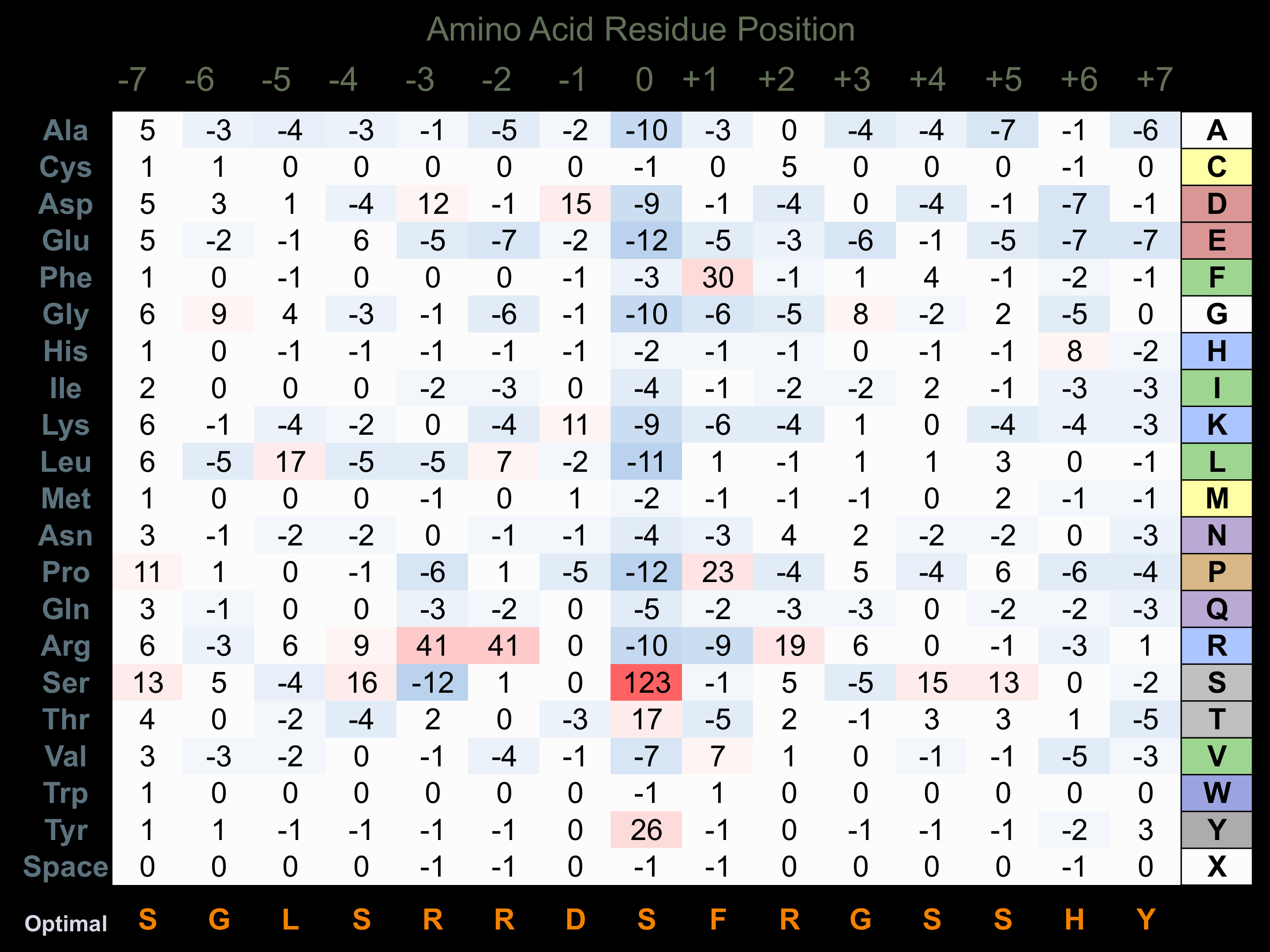
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Cancer, neurological disorders
Specific Diseases (Non-cancerous):
Narcolepsy
Comments:
Nacrolepsy, also known as hypnolepsy, is a neurological disease characterized by the inability of the brain to properly regulate sleep-wake cycles. Symptoms of this disease include excessive daytime sleepiness and cataplexy, which is a sudden onset, transient occurence of muscle weakness with full conscious awareness. Nacrolepsy is throught to result from the autoimmune degeneration of hypocretin/orexin-producing neurons in the brain, which are critical to the regulation of sleep-wake cycles. Elevated levels of antibodies directed against TRIB2 (anti-TRIB2) have been observed in patients with nacrolepsy, indicating that an anti-TRIB2 antibody-mediated degeneration of hypocretin/orexin-producing neurons may contribute to the pathogenesis of nacrolepsy. However, it is unknown as to whether the anti-TRIB2 antibodies cause damage to the hypocretin/orexin-producing neurons.
Specific Cancer Types:
Acute myelogenous leukemias (AML)
Comments:
Trb2 may be an oncoprotein (OP). In animal studies, mice injected with hemapoietic stem cells (HSC) that over-expressed Trb2 displayed a uniform development of acute myelogenous leukemia (AML), indicating a role for Trb2 in the pathogenesis of AML. In addition, Trb2 over-expression is associated with the degradation of C/EBPalpha, a transcription factor that is commonly mutated in patients with AML.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +188, p<0.016); Bladder carcinomas (%CFC= +68, p<0.051); Brain glioblastomas (%CFC= -60, p<0.057); Cervical cancer (%CFC= -64, p<0.0007); Cervical cancer stage 2A (%CFC= -50, p<0.076); Colorectal adenocarcinomas (early onset) (%CFC= +171, p<0.003); Oral squamous cell carcinomas (OSCC) (%CFC= +234, p<0.0001); Ovary adenocarcinomas (%CFC= +130, p<0.003); Prostate cancer (%CFC= -51, p<0.091); Skin fibrosarcomas (%CFC= +752, p<0.003); Skin melanomas - malignant (%CFC= +952, p<0.0001); Uterine leiomyomas (%CFC= -69, p<0.032); and Uterine leiomyomas from fibroids (%CFC= +71, p<0.057). The COSMIC website notes an up-regulated expression score for Trb2 in diverse human cancers of 404, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 3 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.1 % in 24726 diverse cancer specimens. This rate is a modest 1.4-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.6 % in 1270 large intestine cancers tested; 0.58 % in 603 endometrium cancers tested; 0.4 % in 589 stomach cancers tested; 0.27 % in 864 skin cancers tested; 0.25 % in 1634 lung cancers tested; 0.16 % in 710 oesophagus cancers tested; 0.11 % in 548 urinary tract cancers tested; 0.11 % in 273 cervix cancers tested; 0.09 % in 1316 breast cancers tested; 0.07 % in 1276 kidney cancers tested; 0.04 % in 1459 pancreas cancers tested; 0.03 % in 881 prostate cancers tested; 0.03 % in 2082 central nervous system cancers tested; 0.01 % in 2009 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: S515S (7); R268Q (3).
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.

