Nomenclature
Short Name:
ULK3
Full Name:
Serine-threonine-protein kinase ULK3
Alias:
- AL117482
- DKFZP434C131
- Hypothetical protein DKFZp434C131
- Unc-51-like kinase 3
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
ULK
SubFamily:
NA
Structure
Mol. Mass (Da):
53,444
# Amino Acids:
472
# mRNA Isoforms:
4
mRNA Isoforms:
54,997 Da (483 AA; Q6PHR2-4); 53,444 Da (472 AA; Q6PHR2); 53,217 Da (470 AA; Q6PHR2-3); 24,402 Da (214 AA; Q6PHR2-2)
4D Structure:
NA
1D Structure:
Subfamily Alignment
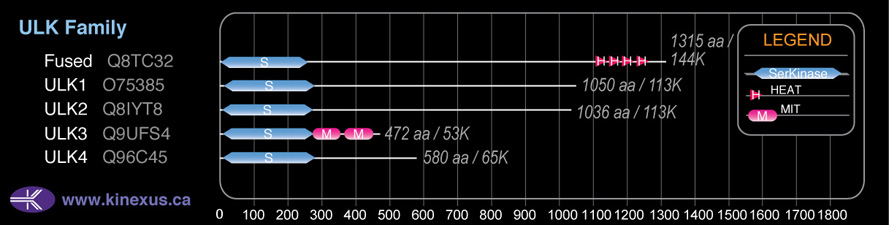
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 74
74
1349
9
896
 4
4
69
1
0
 -
-
-
-
-
 17
17
318
35
405
 51
51
938
10
476
 5
5
94
9
19
 5
5
88
10
34
 100
100
1832
10
3621
 -
-
-
-
-
 16
16
288
32
324
 13
13
231
8
166
 54
54
990
25
503
 -
-
-
-
-
 -
-
-
-
-
 21
21
391
8
308
 5
5
95
5
36
 15
15
267
205
278
 -
-
-
-
-
 8
8
141
25
92
 61
61
1122
28
484
 28
28
506
8
439
 39
39
722
8
567
 -
-
-
-
-
 -
-
-
-
-
 29
29
534
8
415
 85
85
1566
19
1475
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 8
8
155
14
100
 -
-
-
-
-
 79
79
1439
11
2336
 81
81
1493
43
2273
 45
45
821
26
682
 49
49
895
13
406
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.1
99.1
99.5
99 73.7
73.7
77.9
- -
-
-
97 -
-
-
- 75.3
75.3
79
95 -
-
-
- 94
94
97.6
94 93.6
93.6
96.4
95 -
-
-
- -
-
-
- 78.1
78.1
90
79 72.8
72.8
87.2
74 71.1
71.1
84.7
71 -
-
-
- 40.1
40.1
57.6
46 44.1
44.1
64.3
- 23.8
23.8
37.1
- 48
48
64.5
- -
-
-
- -
-
-
- -
-
-
- 26.7
26.7
41.6
- 24.8
24.8
36.3
- 23.8
23.8
35
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
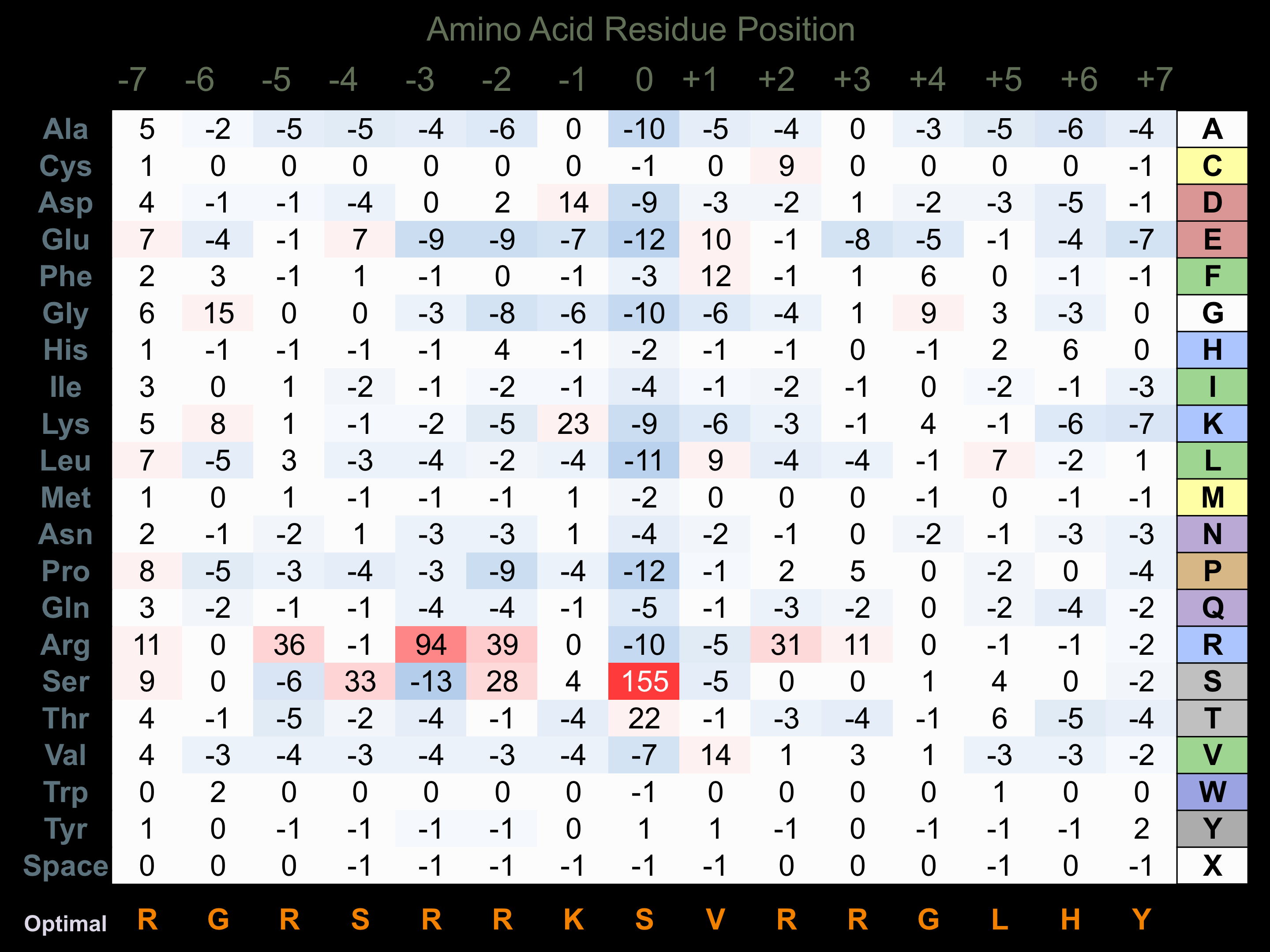
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Chronic myeloid leukemias (CML)
Comments:
ULK3 attenuates positive regulation in cancer and multipotent progenitor cells. ULK3 was identified as a drug target for colon cancer.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Ovary adenocarcinomas (%CFC= +109, p<0.048); Prostate cancer (%CFC= +48, p<0.088); and Prostate cancer - metastatic (%CFC= +54, p<0.015).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. K44R and K139R mutations in ULK3 can lead to reduced phosphotransferase activity.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 24433 diverse cancer specimens. This rate is only -31 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.19 % in 1229 large intestine cancers tested; 0.18 % in 603 endometrium cancers tested; 0.17 % in 864 skin cancers tested; 0.14 % in 881 prostate cancers tested; 0.11 % in 555 stomach cancers tested; 0.08 % in 807 ovary cancers tested; 0.08 % in 1253 kidney cancers tested; 0.07 % in 1437 pancreas cancers tested; 0.07 % in 1289 breast cancers tested; 0.06 % in 382 soft tissue cancers tested; 0.05 % in 1608 lung cancers tested; 0.04 % in 558 thyroid cancers tested; 0.04 % in 548 urinary tract cancers tested; 0.03 % in 2030 central nervous system cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: M180T (3).
Comments:
Only 2 deletions, 3 insertions, and no complex mutations are noted on the COSMIC website.

