Nomenclature
Short Name:
VRK2
Full Name:
Serine-threonine-protein kinase VRK2
Alias:
- EC 2.7.11.1
- Serine/threonine protein kinase VRK2
- Vaccinia related kinase 2
- Vaccinia-related kinase 2
Classification
Type:
Protein-serine/threonine kinase
Group:
CK1
Family:
VRK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
58141
# Amino Acids:
508
# mRNA Isoforms:
5
mRNA Isoforms:
58,141 Da (508 AA; Q86Y07); 55,446 Da (485 AA; Q86Y07-3); 45,034 Da (396 AA; Q86Y07-5); 45,030 Da (397 AA; Q86Y07-2); 44,411 Da (390 AA; Q86Y07-4)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
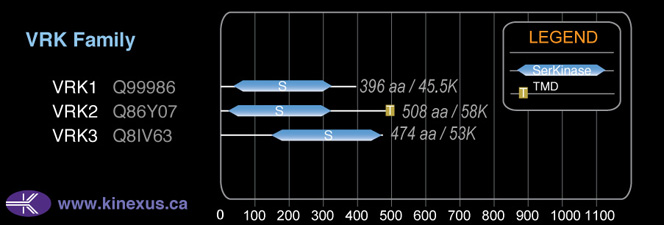
Domain Distribution:
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K93.
Serine phosphorylated:
S339, S369, S406, S438, S445, S447, S449.
Threonine phosphorylated:
T336.
Tyrosine phosphorylated:
Y44.
Ubiquitinated:
K33, K75, K177, K223, K255, K264, K328, K383, K402, K443.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 59
59
1195
22
1136
 3
3
51
9
25
 50
50
1001
13
862
 26
26
521
83
1254
 35
35
696
24
617
 7
7
144
52
145
 11
11
226
29
408
 100
100
2015
35
3248
 20
20
400
10
372
 8
8
163
78
410
 31
31
624
24
1182
 34
34
691
123
800
 42
42
854
24
1487
 3
3
65
9
46
 50
50
1003
22
1952
 4
4
78
13
61
 37
37
750
118
3670
 78
78
1571
18
3764
 16
16
317
73
571
 33
33
666
84
587
 33
33
661
20
789
 50
50
1000
22
1498
 40
40
796
22
1195
 45
45
916
20
1127
 32
32
636
22
856
 42
42
848
60
1057
 42
42
845
27
1300
 51
51
1023
19
1936
 54
54
1082
19
1595
 6
6
111
28
112
 37
37
755
18
490
 25
25
503
26
656
 12
12
239
65
495
 57
57
1143
57
707
 5
5
104
35
147
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 95.6
95.6
96.2
100 87.5
87.5
91.9
97 -
-
-
83.5 -
-
-
85 70.9
70.9
78.3
78.5 -
-
-
- 69.4
69.4
79.9
70 21.8
21.8
35.4
82 -
-
-
- 41.9
41.9
55.3
- 24.7
24.7
39.7
57 26.5
26.5
41.3
48 40.1
40.1
55.7
63 -
-
-
- 30.8
30.8
45.9
- -
-
-
- -
-
-
- 37.4
37.4
52.7
- -
-
-
- -
-
-
- -
-
-
- 21.4
21.4
38.1
- 23
23
40.7
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | RAN - P62826 |
| 2 | TAB1 - Q15750 |
| 3 | MAPK8IP1 - Q9UQF2 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
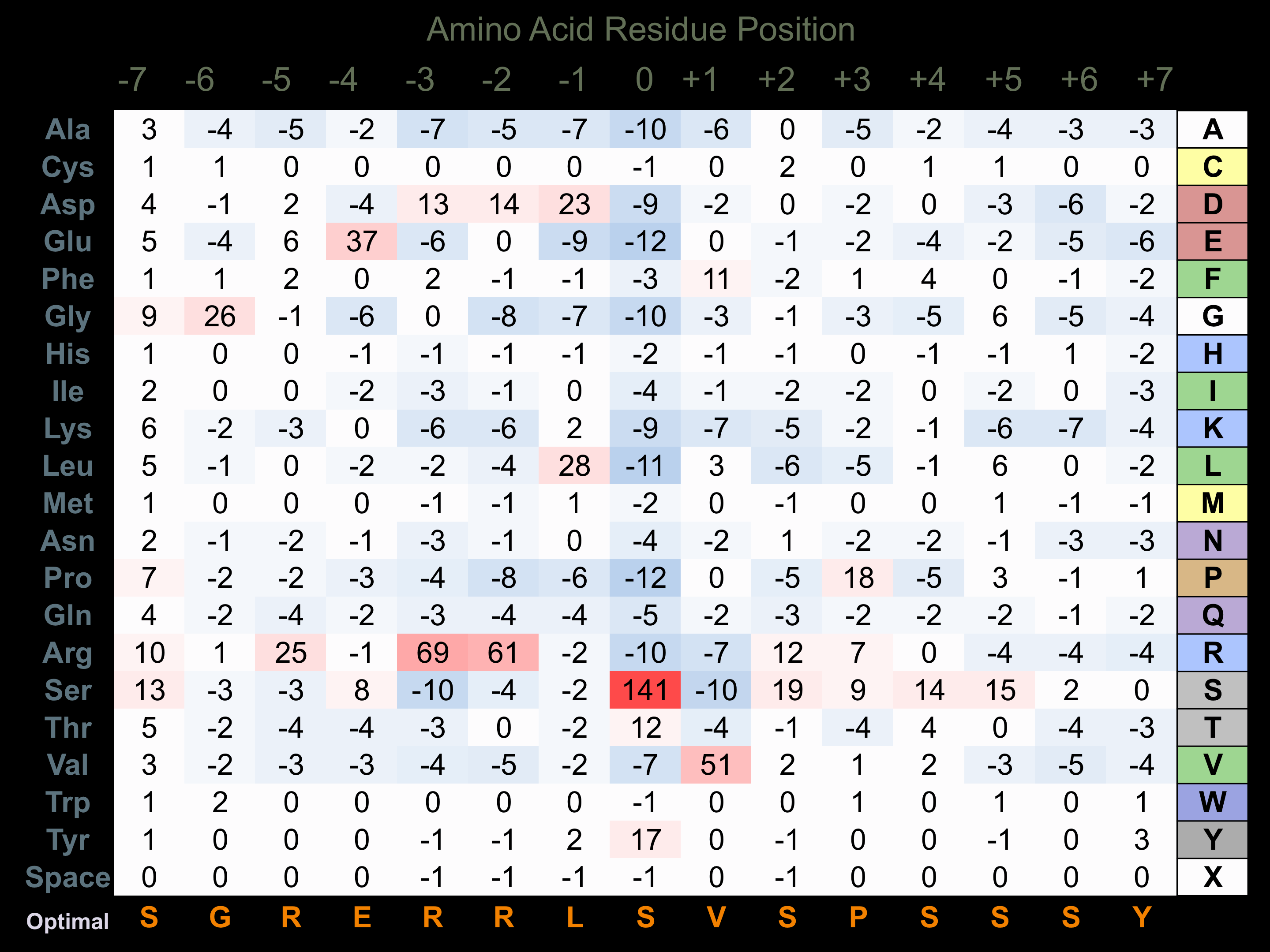
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| IC261 | IC50 > 15 nM | 3674 | 576349 | 21829721 |
| AZD7762 | IC50 = 30 nM | 11152667 | 21829721 | |
| Ro-31-8220 | IC50 = 34 nM | 5083 | 6291 | 21829721 |
| Roscovitine | IC50 > 55 nM | 160355 | 14762 | 21829721 |
| Alvocidib | Kd = 63 nM | 9910986 | 428690 | 22037378 |
| Bosutinib | Kd = 710 nM | 5328940 | 288441 | 22037378 |
| Aurora A Inhibitor 29 (DF) | Kd = 1 µM | 21992004 | ||
| MK5108 | IC50 > 1 µM | 24748204 | 20053775 | |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| Dasatinib | Kd = 3.2 µM | 11153014 | 1421 | 22037378 |
| A674563 | Kd = 4 µM | 11314340 | 379218 | 22037378 |
| Ruxolitinib | Kd = 4 µM | 25126798 | 1789941 | 22037378 |
Disease Linkage
General Disease Association:
Infectious disease
Specific Diseases (Non-cancerous):
Vaccinia; Schizophrenia; Benign familial infantile epilepsy
Comments:
The VRK2 gene has been mapped to chromosome 2p16-p15 and mutations in this gene have been associated with schizophrenia. In a recent genome wide association study (GWAS), a common structural variant of VRK2 was found as a genetic factor with strong association to schizophrenia, along with NRGN, TCF4, and CCDC68. Vaccinia is a viral infection of the skin that is due to the vaccinia virus, which is used as a vaccine against smallpox. The infection is transmitted through contact with the open vaccination site or through fomites that have been contaminated with the virus from the vaccination site. Symptoms include skin irritation, rash, fever, and body aches.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Classical Hodgkin lymphomas (%CFC= +90, p<0.051); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= -82, p<0.0001); Colon mucosal cell adenomas (%CFC= +98, p<0.0001); and Vulvar intraepithelial neoplasia (%CFC= +62, p<0.079). The COSMIC website notes an up-regulated expression score for VRK2 in diverse human cancers of 340, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 88 for this protein kinase in human cancers was 1.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24728 diverse cancer specimens. This rate is only -6 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.37 % in 1270 large intestine cancers tested; 0.29 % in 603 endometrium cancers tested; 0.22 % in 1636 lung cancers tested; 0.18 % in 864 skin cancers tested; 0.17 % in 589 stomach cancers tested; 0.14 % in 710 oesophagus cancers tested; 0.08 % in 1512 liver cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R5I (3); .
Comments:
Only 7 deletions (6 at V364fs*21), 2 insertions, and no complex mutations are noted in COSMIC website.

