Nomenclature
Short Name:
WEE2
Full Name:
Wee1-like protein kinase 1B
Alias:
- FLJ16107
- WEE1 2
- Wee2
- WEE1B
- Wee1b
- Wee1B kinase
- Wee1-like protein kinase 1B
Classification
Type:
Dual specificity protein kinase
Group:
Other
Family:
WEE
SubFamily:
NA
Structure
Mol. Mass (Da):
62,925
# Amino Acids:
567
# mRNA Isoforms:
1
mRNA Isoforms:
62,925 Da (567 AA; P0C1S8)
4D Structure:
NA
1D Structure:
Subfamily Alignment
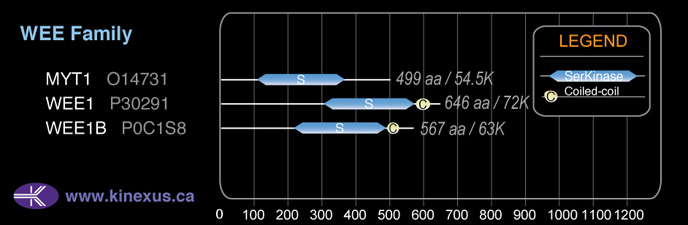
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 212 | 490 | Pkinase |
| 494 | 519 | Coiled-coil |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S102.
Threonine phosphorylated:
T83, T225, T509.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 -
-
-
-
-
 15
15
494
2
552
 -
-
-
-
-
 100
100
3273
5
5557
 -
-
-
-
-

 -
-
-
-
-
 13
13
428
4
464
 -
-
-
-
-
 21
21
678
4
707
 17
17
556
4
581
 14
14
452
4
476
 7
7
233
2
1
 88
88
2877
2
3078
 15
15
493
4
599
 27
27
892
2
502
 2
2
54
79
87
 38
38
1251
2
343
 76
76
2479
2
798
 98
98
3220
2
397
 8
8
268
4
237
 1.3
1.3
41
2
2
 -
-
-
-
-
 46
46
1498
4
870
 8
8
252
4
388
 25
25
807
2
61
 19
19
625
2
182
 33
33
1093
2
209
 41
41
1350
2
666
 -
-
-
-
-
 -
-
-
-
-
 0.7
0.7
24
8
45
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 77.7
77.7
78.1
100 94.3
94.3
95.9
94.5 -
-
-
74 -
-
-
- 71.9
71.9
77.7
79 -
-
-
- 66.8
66.8
76.5
69 43
43
56.8
68 -
-
-
- 47.7
47.7
57.1
- 42.1
42.1
57.8
57 52.9
52.9
64.9
52 43.5
43.5
58.9
54.5 -
-
-
- 33.5
33.5
50.7
- 36.1
36.1
52.6
- -
-
-
- 39.2
39.2
55.2
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
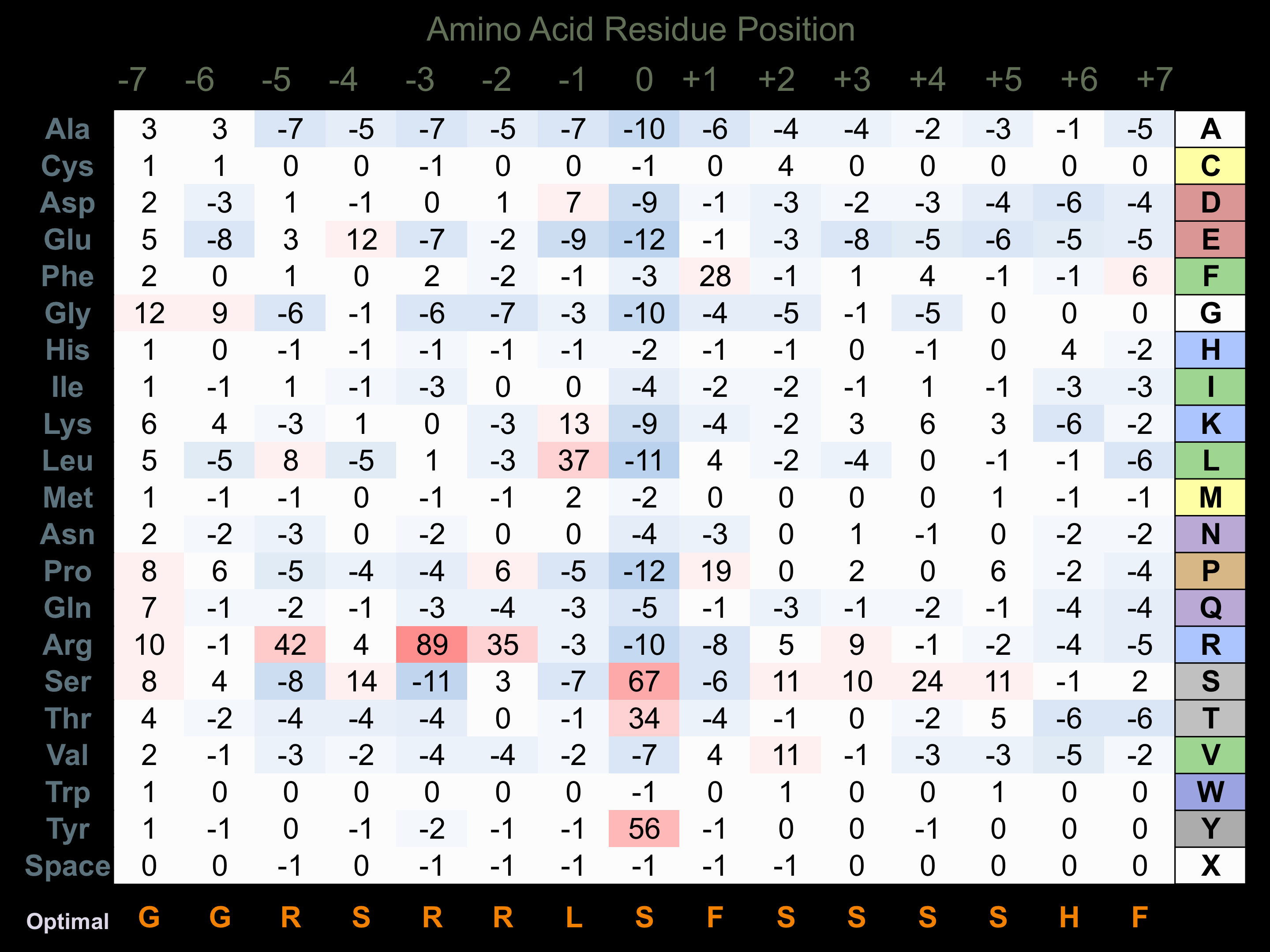
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| PD173955 | Kd = 57 nM | 447077 | 386051 | 22037378 |
| Bosutinib | Kd = 94 nM | 5328940 | 288441 | 22037378 |
| Dasatinib | Kd = 200 nM | 11153014 | 1421 | 22037378 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| Neratinib | Kd = 1.3 µM | 9915743 | 180022 | 22037378 |
| Tozasertib | Kd = 4.2 µM | 5494449 | 572878 | 22037378 |
| KW2449 | Kd = 4.7 µM | 11427553 | 1908397 | 22037378 |
Disease Linkage
General Disease Association:
Infectious disease
Specific Diseases (Non-cancerous):
Western equine encephalitis
Comments:
Western equine encephalitis is a viral infectious diease characterized by the presence of inflammation in the brain, which can affect both horses and humans. The causitive agent of the disease is the Western equine encephalomyelitis virus, which can be transmitted by mosquitoes of the genera Culex and Aedes. Symptoms of the infection include fever, headache, nausea, vomiting, anorexia, altered mental status, and overall weakness.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= -80); and Brain oligodendrogliomas (%CFC= -79). The COSMIC website notes an up-regulated expression score for WEE2 in diverse human cancers of 431, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 24698 diverse cancer specimens. This rate is only 23 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.57 % in 864 skin cancers tested; 0.41 % in 1259 large intestine cancers tested; 0.35 % in 603 endometrium cancers tested; 0.22 % in 629 stomach cancers tested; 0.16 % in 548 urinary tract cancers tested; 0.15 % in 238 bone cancers tested; 0.15 % in 1634 lung cancers tested; 0.1 % in 710 oesophagus cancers tested; 0.09 % in 382 soft tissue cancers tested; 0.06 % in 273 cervix cancers tested; 0.06 % in 1512 liver cancers tested; 0.04 % in 942 upper aerodigestive tract cancers tested; 0.04 % in 881 prostate cancers tested; 0.04 % in 833 ovary cancers tested; 0.04 % in 1316 breast cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R242C (3); R410W (3).
Comments:
Only 3 deletions, and no insertions or complex mutations are noted on the COSMIC website.

