Nomenclature
Short Name:
YSK1
Full Name:
Serine-threonine-protein kinase 25
Alias:
- DKFZp686J1430
- Ste20-like kinase
- Sterile 20,oxidant stress-response kinase 1
- STK25
- EC 2.7.11.1
- SOK1
- ST25
- Ste20,oxidant stress response kinase-1
Classification
Type:
Protein-serine/threonine kinase
Group:
STE
Family:
STE20
SubFamily:
YSK
Specific Links
Structure
Mol. Mass (Da):
48,112
# Amino Acids:
426
# mRNA Isoforms:
3
mRNA Isoforms:
48,112 Da (426 AA; O00506); 39,266 Da (349 AA; O00506-2); 37,297 Da (332 AA; O00506-3)
4D Structure:
Homodimer.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
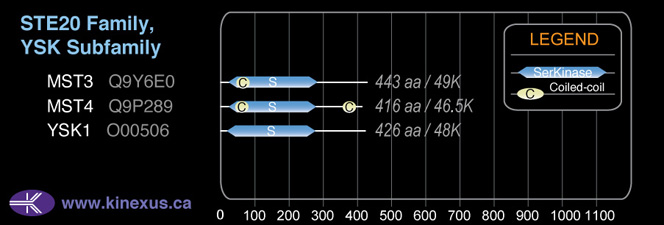
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 20 | 270 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S12, S30, S231, S278, S342.
Threonine phosphorylated:
T166+, T168+, T174+, T178-, T360.
Tyrosine phosphorylated:
Y35.
Ubiquitinated:
K142, K171.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 25
25
841
16
953
 2
2
53
11
22
 4
4
144
1
0
 80
80
2700
182
5577
 21
21
717
14
666
 25
25
832
51
2071
 15
15
513
23
652
 39
39
1309
27
2225
 9
9
301
10
247
 5
5
163
58
117
 75
75
2534
112
5790
 17
17
582
118
632
 3
3
89
12
16
 2
2
56
11
52
 4
4
118
12
141
 4
4
134
9
29
 9
9
309
186
2470
 2
2
63
8
34
 96
96
3224
198
6410
 17
17
578
56
595
 4
4
130
14
147
 4
4
137
16
151
 10
10
335
10
137
 100
100
3358
138
6095
 6
6
194
12
219
 30
30
1015
34
1383
 2
2
74
15
32
 2
2
53
9
42
 3
3
93
10
35
 1.2
1.2
39
14
12
 36
36
1216
18
799
 25
25
837
21
1032
 0.4
0.4
13
17
10
 20
20
681
31
615
 6
6
215
22
70
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 72.4
72.4
72.6
99.5 38.4
38.4
57.1
99 -
-
-
95 -
-
-
98 71.6
71.6
72.5
97 -
-
-
- 98.6
98.6
99.8
99 38.5
38.5
55.2
99 -
-
-
- 94.4
94.4
97.9
- 38.9
38.9
56
92 38.5
38.5
56.2
82 38.6
38.6
56.9
83 -
-
-
- 29.8
29.8
42.1
- -
-
-
- 35.6
35.6
51.7
66 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 20.8
20.8
28.3
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PDCD10 - Q9BUL8 |
| 2 | PPP2R1A - P30153 |
| 3 | SLMAP - Q14BN4 |
| 4 | TCP1 - P17987 |
| 5 | STRN - O43815 |
| 6 | PPP2R1B - P30154 |
| 7 | CTTNBP2NL - Q9P2B4 |
| 8 | FAM40A - Q5VSL9 |
| 9 | SIKE1 - Q9BRV8 |
| 10 | FAM40B - Q9ULQ0 |
| 11 | PPP2CA - P67775 |
| 12 | MOBKL3 - Q9Y3A3 |
| 13 | STRN4 - Q9NRL3 |
| 14 | FGFR1OP2 - Q9NVK5 |
| 15 | CCM2 - Q9BSQ5 |
Regulation
Activation:
NA
Inhibition:
The C-terminal non-catalytic region inhibits the kinase phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
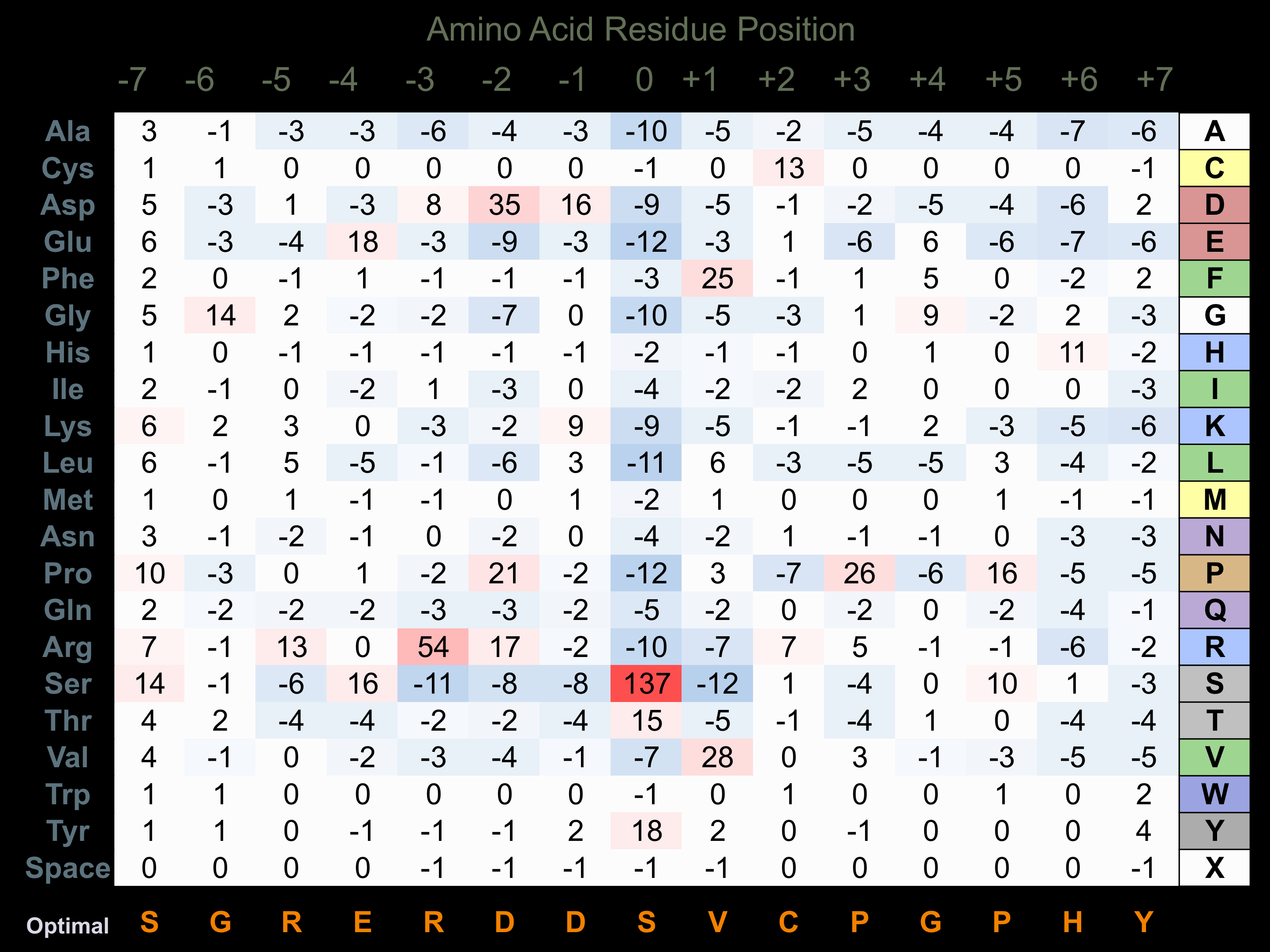
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Genetic disorders
Specific Diseases (Non-cancerous):
Anoxia; Pseudopseudohypoparathyroidism
Comments:
Pseudopseudohypoparathyroidism (PPHP) is an inherited endocrine disease characterized by a suite of clinical features referred to as the albright hereditary osteodystrophy (AHO) phenotype, including short stature, obesity, round face, short hand bones, and in some cases, intellectual disability. Affected individuals are not resistant to parathyroid hormone (PTH), thus they do not have hypoparathyroidism. Cellular YSK1 activity increases 3-7 fold after exposure to reactive oxygen intermediates and mutations in the kinase are associated with an increased occurence of cerebral hypoxia (a lack of oxygen to the brain). The mouse YSK1 gene has been mapped to the central region of chromosome 1, which shares homology with the long arm of chromosome 2 in humans. Therefore, it is predicted that the human YSK1 gene would map to this region of chromosome 2 (specifically 2q37). Deletion mutations of the 2q37 chromosome region have been implicated in the pathogenesis of PPHP. Genomic characterization of a subset of PPHP patients reveled the presence of several small deletion mutations near the distal end of chromosome 2, in the vicinity of the 2q37 region. Furthermore, DNA samples from this PHPP patient subset showed no hybridization to a YSK1 gene probe, indicating that the gene had been deleted. Thus, loss-of-function mutations in YSK1 have been implicated in the pathogenesis of PPHP.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Breast epithelial cell carcinomas (%CFC= +73, p<0.004); Large B-cell lymphomas (%CFC= +48, p<0.003); Malignant pleural mesotheliomas (MPM) tumours (%CFC= +85, p<0.009); and T-cell prolymphocytic leukemia (%CFC= +65, p<0.074). The COSMIC website notes an up-regulated expression score for YSK1 in diverse human cancers of 525, which is 1.1-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 288 for this protein kinase in human cancers was 4.8-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 25371 diverse cancer specimens. This rate is only -24 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.49 % in 1093 large intestine cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,654 cancer specimens
Comments:
Only 2 deletions, and no insertions or complex mutations are noted on the COSMIC website.

